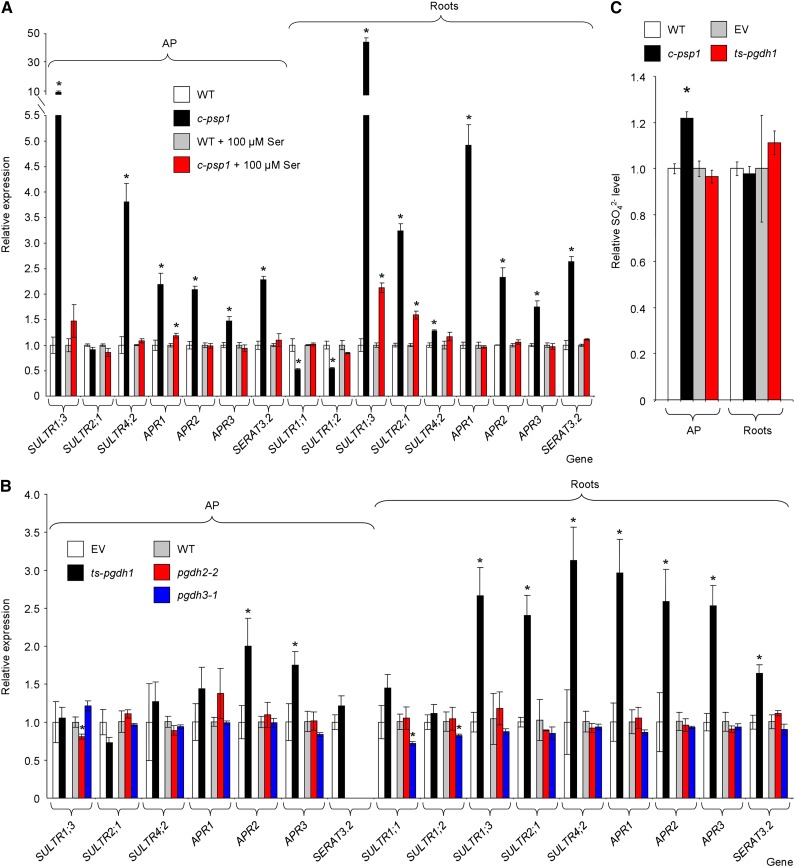
Figure 4.
Expression of sulfur metabolism-related genes in PPSB-deficient lines during the daytime. A, qRT-PCR expression analysis in c-psp1 in the presence or absence of added l-Ser. B, qRT-PCR expression analysis in PGDH family deficient lines (ts-pgdh1, pgdh2-2, and pgdh3-1). C, Sulfate content in AP and roots of c-psp1 and ts-pgdh1. Data for c-psp1, pgdh2-2, and pgdh3-1 are relative values normalized to the mean calculated for wild-type plants (WT). Data for ts-pgdh1 lines were normalized to the values of the wild type transformed with the EV. Samples were collected in the middle of the light period. Values represent the means ± se; n > 3 pools of 30 plants. Both c-psp1 and ts-pgdh1 material proceeded from pools of two different transgenic events. *Significantly different to control lines (P < 0.05, Student’s t test).