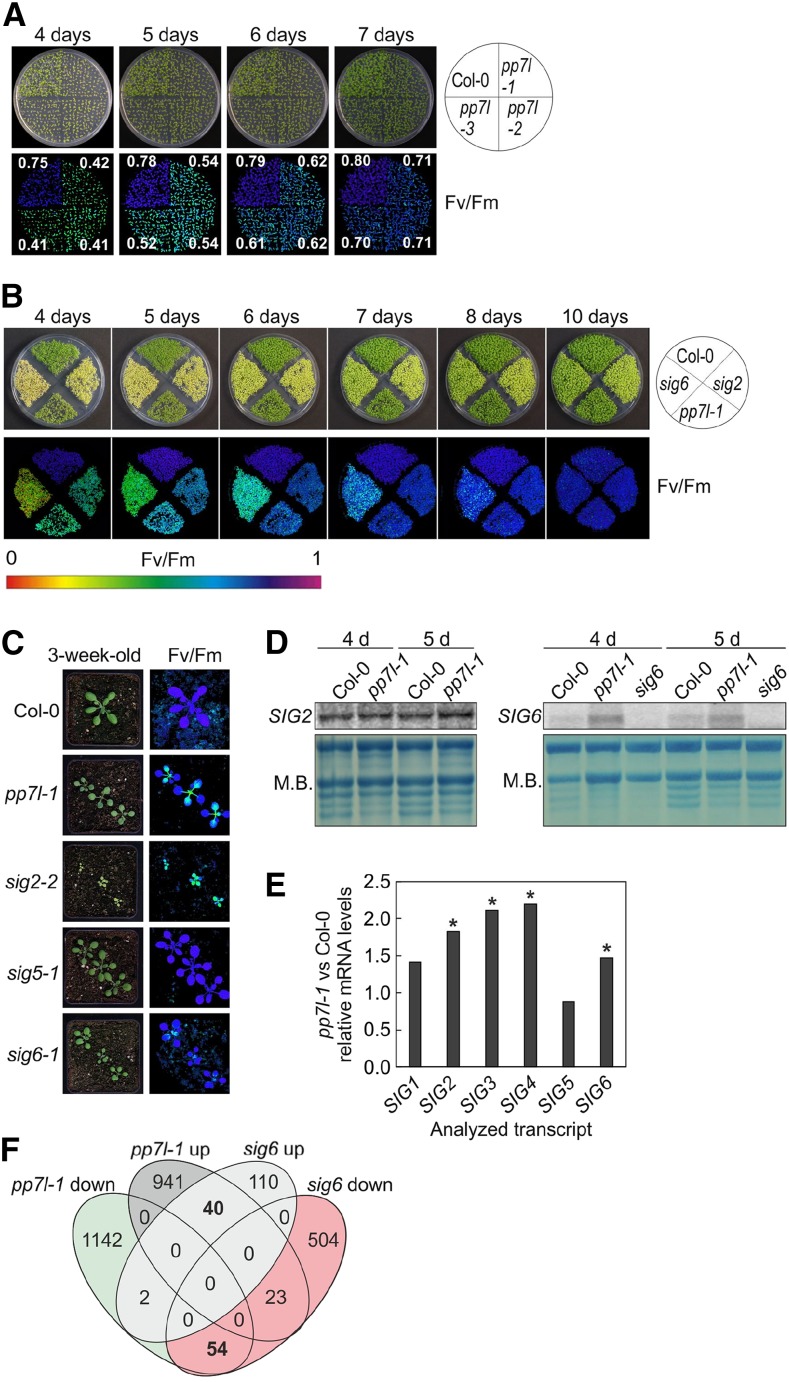
Figure 3.
Comparative analysis of pp7l-1 and sig (sig2 and sig6) mutants. A, Phenotypes of 4- to 7-d–old Col-0 and the different pp7l mutant seedlings together with Imaging PAM pictures showing maximum quantum yields of PSII (Fv/Fm) sds are provided in Supplemental Table S1. B, Phenotypes and Imaging PAM images representing the Fv/Fm activity of 4- to 10-d–old Col-0, pp7l-1 and sig2 and sig6 mutant seedlings. C, Phenotypes of 3-week–old Col-0, pp7l-1, and sig mutant plants and Imaging PAM pictures showing maximum quantum yields of PSII (Fv/Fm). Scale bars = 1 cm. D, Northern-blot analysis of SIG2 and SIG6 transcripts in 4- and 5-d–old Col-0 and pp7l-1 plants. For the detection of SIG6 mRNA levels, the sig6 mutant was included as a control, because several unspecific bands were detected with the SIG6 probe. Total RNAs were fractionated in a formaldehyde-containing denaturing gel, transferred onto a nylon membrane, and probed with (α-32P)dCTP-labeled cDNA fragments specific for the different SIG transcripts. The rRNA was visualized by staining the membrane with methylene blue (M.B.) and served as a loading control. E, Graph representing the changes (relative to Col-0 values) in SIG mRNA levels in pp7l-1, as determined by lncRNA-Seq analysis. Significant differences (DESeq2 analysis running with the fit type set to “parametric”; adjusted P < 0.05) with respect to Col-0 are denoted by asterisks. The sd was not calculated, because differentially expressed genes were identified with “parametric” and not “mean” fit settings in DESeq2. F, Venn diagrams depicting the degree of overlap between the sets of genes whose expression levels were altered by at least twofold (up or down) in the pp7l‐1 mutant compared with the sig6 mutant.