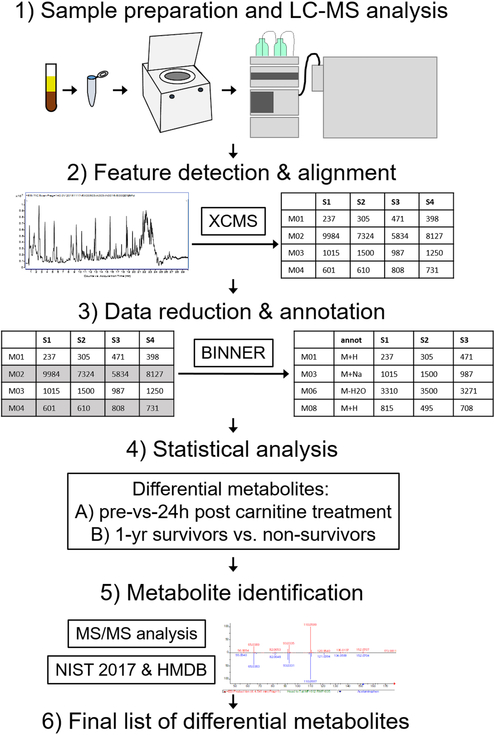
Figure 1:
Schematic of the sample analysis and data processing workflow. Serum samples were extracted with solvent and subjected to LC-MS analysis (step 1). Features were detected and aligned in the resulting data using XCMS (step 2). Data were then processed using data reduction and annotation software Binner, which enabled removal of redundant features including isotopes, fragments and minor adducts while annotating “primary” features with putative ion type (M+H, M+Na, etc) (step 3). Remaining features were subject to univariate statistical analysis with FDR correction to detect differential features between pre and 24h post carnitine samples, as well as between 1-year sepsis survivors and non-survivors (step 4). Differential features were subject to MS/MS analysis and database search to assign metabolite identity when possible or provide putative annotation when not (step 5), resulting in the final lists of differential metabolites included in this manuscript.