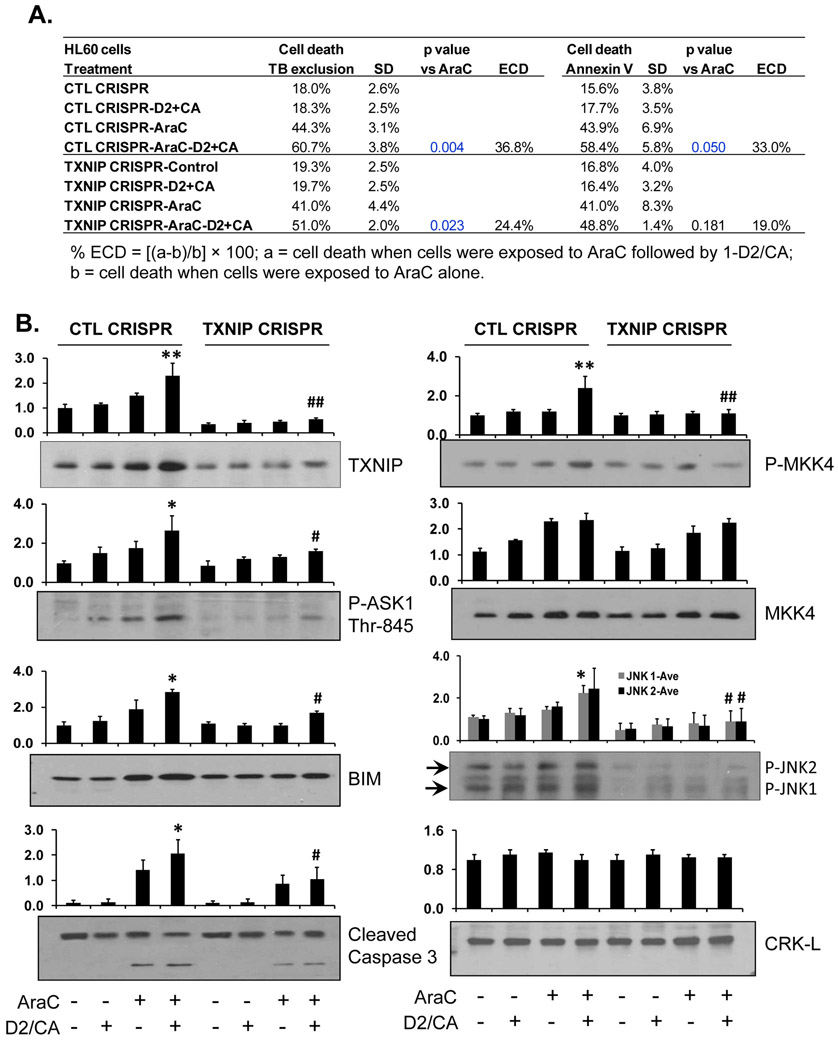
Figure 3. Knock down of TXNIP by CRISPR/Cas9 in HL60 cells reduces ECD.
A. Depletion of TXNIP by CRISPR/Cas9 in HL60. The pretreatment with AraC for 72 h, then addition of D2/CA for 96 h. The percentages of cell death, as determined by trypan blue exclusion or annexin V/PI staining, are shown in the table. B. Western blotting to detect the TXNIP downstream targets, ASK1, MKK4 and JNK, as well as the apoptosis-related proteins, BIM and caspase 3. CRK-L was used as a loading control. The average of relative signal intensities from three separate experiments are shown in bar charts above each blot. *, p<0.05; **, p < 0.01 vs the AraC group; #, p<0.01 when comparing CTL CRISPR-ECD with VDR CRISPR-ECD.