Figure 3.
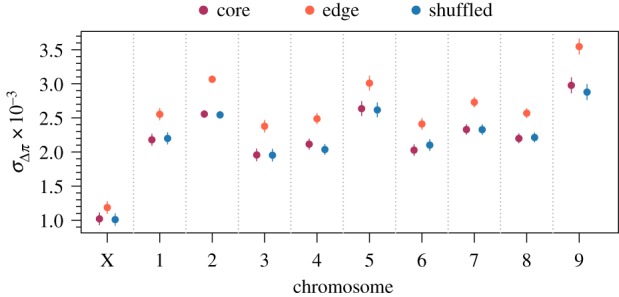
Model estimated standard deviation of changes in nucleotide diversity among replicates at individual windows in different chromosomes along the genome. Higher values of indicate a larger role for neutral processes like gene surfing. Estimates were generated from a linear mixed model with fixed effects for spatial structure, chromosome, and their interaction and random effects capturing the spatial autocorrelation of individual windows along the genome. Only windows with complete sets of observations across all replicates were used for this analysis (n = 6602 windows for each level of spatial structure). Points are model estimates and error bars are 95% confidence intervals. Likelihood ratio tests found all fixed effects significant with p < 0.0001.
