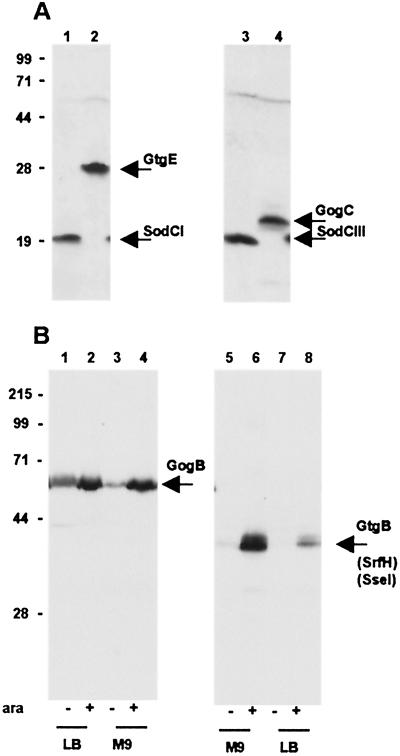
Figure 2.
Immunodetection of epitope-tagged proteins from Salmonella prophages. Whole-cell bacterial lysates were subjected to electrophoretic separation in a 12% polyacrylamide-SDS gel. Proteins were transferred onto poly(vinylidene difluoride) membrane and probed with anti-FLAG M2 mAb (Sigma). (A) Bacteria grown in LB medium: 1, strain MA7172 (sodCI-30∷3xFLAG KnR); 2, strain MA7180 (gtgE-34∷3xFLAG KnR); 3, strain MA7191 (sodCIII-35∷3xFLAG KnR); and 4, strain MA7192 (gogC-36∷3xFLAG KnR). Expected molecular masses for the 3xFLAG fusion proteins are: SodCI and SodCIII, 19 kDa; GtgE, 29 kDa; and GogC, 22 kDa. (B) Bacteria grown under conditions eliciting SsrB-mediated regulation. Lanes 1–4: strain MA7184 (gogB-33∷3xFLAG KnR ssrB∷ Cm/pBAD ssrB+) grown in LB medium (lane 1), in LB medium supplemented with arabinose (lane 2), in M9-glucose medium (lane 3), and in M9-arabinose medium (lane 4). Lanes 5–8: strain MA7185 (gtgB-37∷3xFLAG KnR ssrB∷Cm/pBAD ssrB+) grown in M9-glucose medium (lane 5), in M9-arabinose medium (lane 6), in LB medium (lane 7), and in LB medium supplemented with arabinose (lane 8). Expected molecular masses for the 3xFLAG fusion proteins are: GtgB, 38 kDa and GogB, 57 kDa. Positions and molecular masses (kDa) of protein standards are indicated on the left.