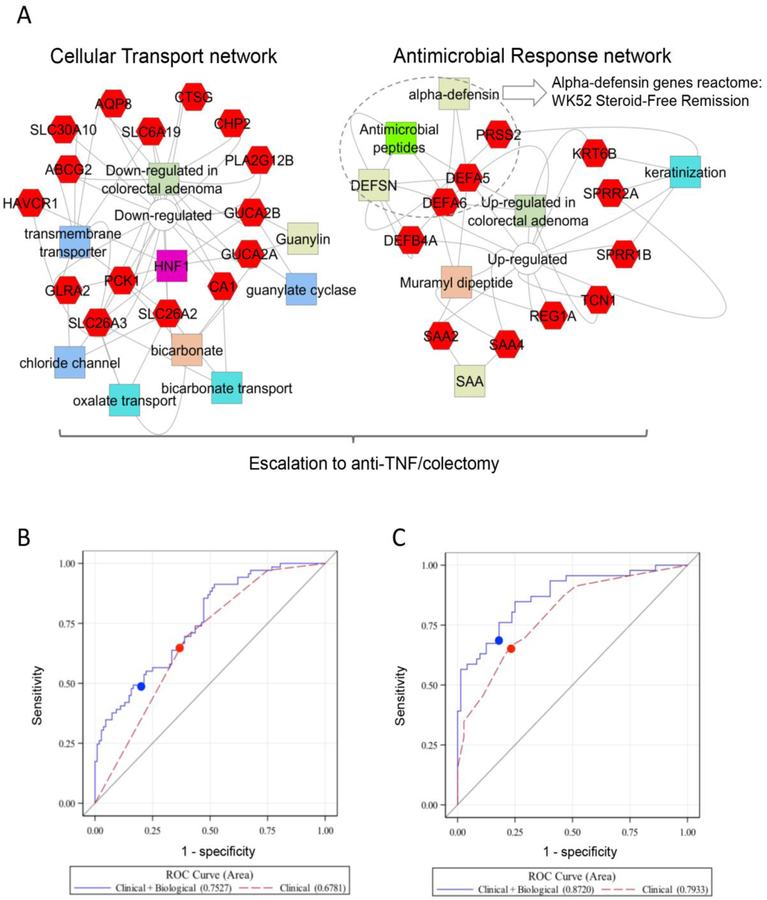
Figure 2. Biological Predictors of Disease Outcome.
A. Gene set enrichment analyses (GSEA) of the 33 genes differentially expressed in the rectum [18 increased (left), and 15 decreased (right)] in those that did vs. did not achieve WK52 CS-free remission using ToppGene, ToppCluster, and Cytoscape are shown (See Supplemental Appendix). The increased gene sets were associated with cellular transport and channel functions, while the decreased gene sets were associated with antimicrobial responses. The overall 33 gene set signature was associated with need for escalation to anti-TNFα, while the alpha-defensins reactome (biosystem ID: 1269266) antimicrobial peptides pathway showed a stronger association with the W52 SFR outcome. Genes are indicated in red hexagons, GO annotations in blue colors, drug-associations in orange, and pathways, gene family, interactions, and co-expression in green colors. The full list of GSEA results and p-values are in Table S4. B and C. ROC curves are presented for the logistic regression models in patients with clinical data (red) and clinical+biological data (blue) from the imputed dataset with the median CV AUC for the models for B) CS-free Remission and C) Escalation to Anti-TNFα. The cut-point used to define sensitivity and specificity for each model is indicated by the filled circle. See Table 2 and Table 3 for the AUCs and the p-value comparing the AUC of the two ROC curves averaged across the multiple imputations.