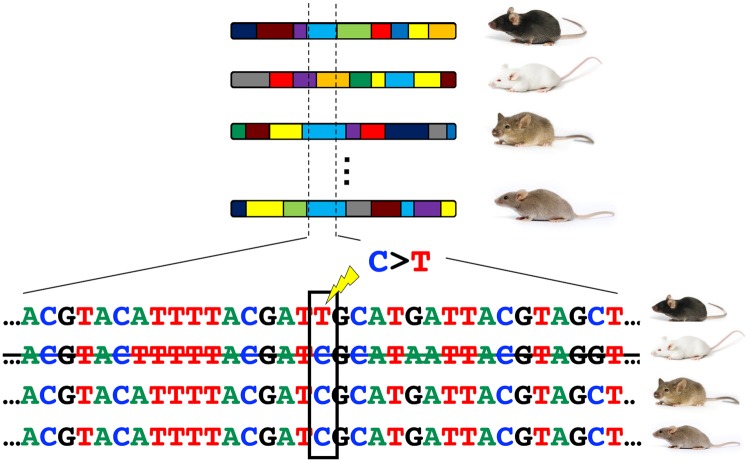
Fig. 1.
Schematic of the approach used to identify putative de novo mutations in inbred laboratory mouse strains. The genomes of laboratory mice can be envisaged as mosaics of less than ten haplotypes derived from a small population of founder animals. The ancestral haplotype structure of four strains is illustrated, with unique haplotypes depicted in different colors. Given the small number of founders, multiple strains are likely to share the same haplotype at a given locus by virtue of their descent from a common ancestor. An example of one such region shared by three of the four depicted strains (light blue haplotype) is outlined by dashed lines. Recent mutations that arose in a single focal strain can be detected as strain-private variants resident on haplotypes that are shared identical-by-descent between strains.