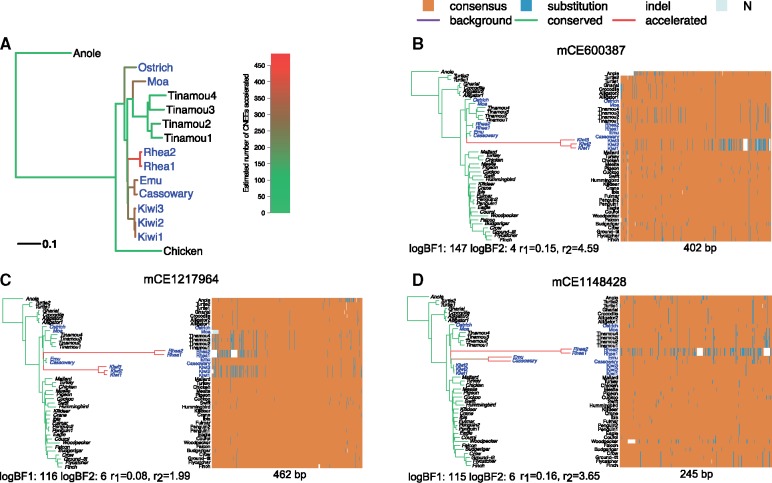
Fig. 4.
(A) Number of accelerated elements per branch among ratite-specific accelerated CNEEs. Phylogeny for avian data set (only a subset of species are shown for illustration). Palaeognaths consist of the flightless ratites and volant tinamous. Ratites are shown in blue. Branch lengths represent the background substitution rates. The gradient of the color indicates the expected number of elements being accelerated under the full model on that branch among 786 ratite-specific accelerated CNEEs. (B–D) Examples of ratite-accelerated CNEEs. For each element, the shift pattern of substitution rates under the full model is shown on the left represented by a phylogenetic tree with branch lengths proportional to the posterior mean of the substitution rate and colored by the posterior mean of Z (green is the conserved, red is the accelerated, and purple is the background state). Longer and redder branch indicates acceleration occurred at a higher rate or earlier on the branch, whereas shorter and greener one means later on the branch or no acceleration. Below the tree shows two log-BFs and conserved ()/accelerated rate (). In the sequence alignment heatmap on the right, each column is one position, each row is a species, and the element length is shown below. For each position, the majority nucleotide (T, C, G, A) among all species is labeled as “consensus” and colored as orange; others are labeled as “substitution” and colored as blue; unknown sequence is labeled as “N” and colored as gray; indels are shown as white space.