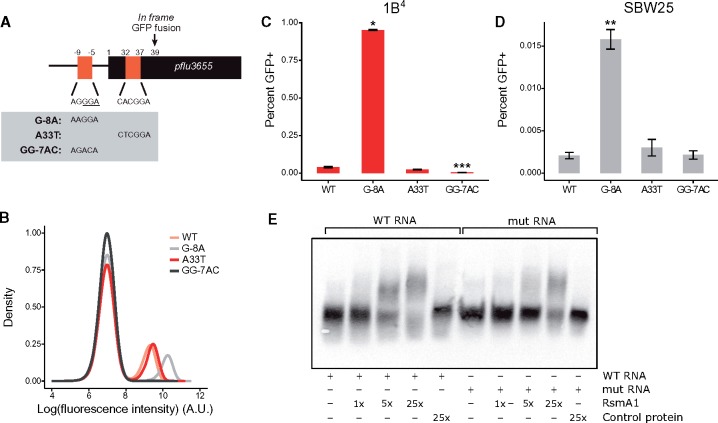
Fig. 4.
RsmA/E binding sites in pflu3655 control capsulation. (A) Schematic diagram of pflu3655 5′-region. Two putative RsmA/E binding sites (orange squares) are located in the promoter and 5′-region of the gene. Numbers indicate nucleotide positions relative to start codon (not to scale). Sequences of putative RsmA/E binding sites are shown, the putative RBS is underlined. Gray box: Point mutations introduced in the different sequences by site-directed mutagenesis. (B) Expression of the Ppflu3655-GFP reporter carrying the different point mutations in the 1B4 background. GFP fluorescence was measured by flow cytometry. Data are representative of three independent experiments. (C, D) Mutations in putative RsmA/E binding sites affect capsulation in 1B4 (C) and SBW25 (D). Individual point mutations were reintroduced into 1B4 and SBW25 carrying the wild-type Ppflu3655-GFP reporter and the proportion of GFP positive cells in late exponential phase (OD600nm = 1–2) was measured by flow cytometry. Means ± SEM are shown, n = 9 (1B4) or n = 7 (SBW25). Data are pooled from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskall–Wallis test with Dunn’s post hoc correction, comparison to 1B4. (E) Specific interaction between RsmA1 and pflu3655 mRNA. EMSA experiments were performed with 6.25 nM biotin-labelled oligonucleotides, either using a wild-type pflu3655 sequence (WT RNA; left) or a version carrying both G-8A and A33T point mutations (mut RNA; right). An increasing concentration of purified His6-RsmA1 (RsmA1) or a high concentration of proteins purified from SBW25 expressing a non-His6-tagged version of RsmA1 (Control protein) was added to the reactions.