Figure 1. Gm26878 deletion analysis.
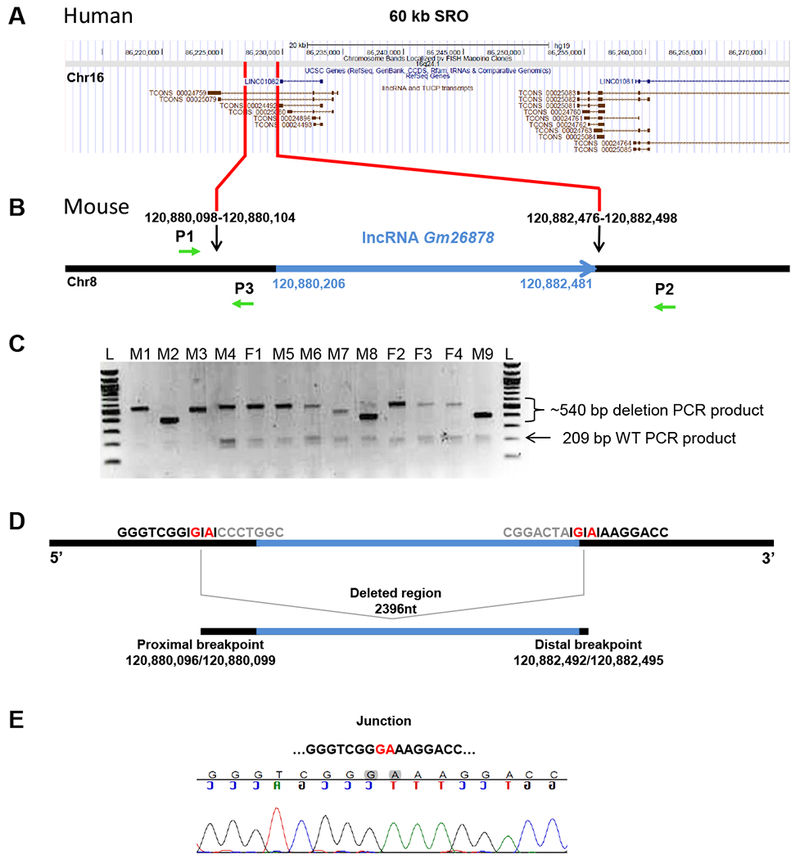
(A) RefSeq annotation (GRCh37/hg19) of the FOXF1 upstream enhancer region SRO (Smallest deletion Region of Overlap delineating the enhancer) of human chromosome 16 corresponding to a fragment of mouse chromosome 8 harboring Gm26878 lncRNA locus. (B) Schematic representation of Gm26878 lncRNA locus (blue) and neighboring sequences (black). Black arrows indicate sgRNA target sites (chr8:120,880,082-120,880,104 and chr8:120,882,476-120,882,498; GRCm38/mm10); green arrows indicate P1, P2, and P3 primers used for genotyping and sequencing (P1 and P3 amplify the wild-type allele; P1 and P2 amplify interval deletion alleles). (C) Agarose gel electrophoresis of products amplified using three PCR primers P1, P2, and P3 on gDNA extracted from 13 founder mice from co-injection of Cas9 and sgRNAs; M = male, F = female, L = ladder, 209 bp = wild-type PCR product, ~540 bp = approximate size of interval deletion PCR products. (D) Deleted region harboring Gm26878 lncRNA with proximal (chr8:120,880,096/120,880,099; GRCm38/mm10) and distal (chr8:120,882,492/120,882,495; GRCm38/mm10) breakpoints. The exact position of breakpoints is unknown due to 2 bp microhomology (marked in red). (E) Chromatogram showing DNA sequences of the nonhomologous end joining (NHEJ) repair junction.
