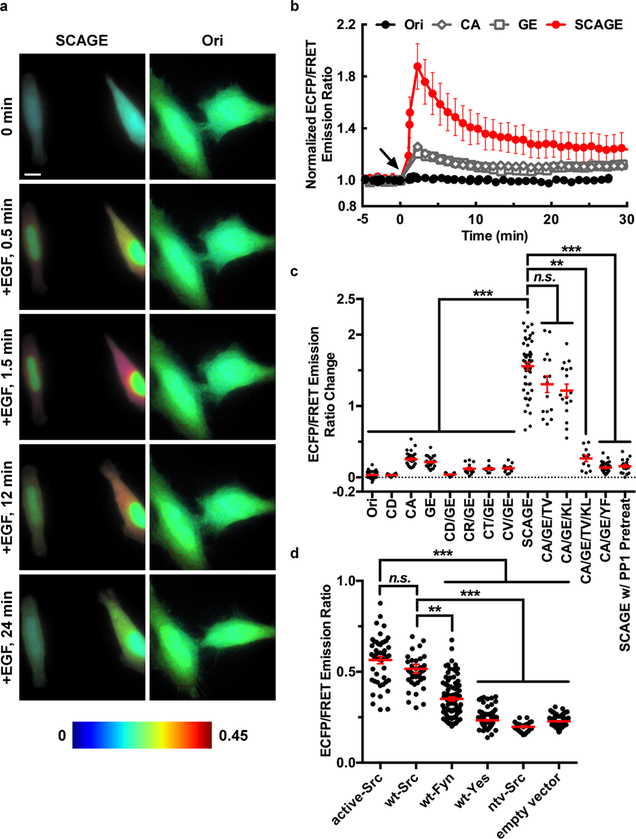
Figure 2.
Characterization of FRET-based Src biosensors in living mammalian cells. (a) Time-lapse FRET imaging in HeLa cells expressing different Src FRET-based biosensors (SCAGE or Ori) stimulated with 50 ng/mL EGF (arrow at 0 min). The color scale at the bottom shows the range of ECFP/FRET emission ratio, with cool and warm colors representing low and high ratios, respectively. Scale bars, 5 μm. (b) Time courses of the normalized ECFP/FRET emission ratio of the Ori (black solid circles, n = 6), CA (open diamonds, n = 7), GE (open squares, n = 8), and SCAGE biosensors (red solid circles, n = 5) in HeLa cells before and after 50 ng/mL EGF stimulation. The ECFP/FRET emission ratios were normalized against the averaged values before EGF stimulation. The error bars represent SEM (the standard error of the mean) and average data are presented as mean ± SEM. (c) Responses of different Src FRET-based biosensors in HeLa cells stimulated with 50 ng/mL EGF. The CA/GE/YF group is the inactive control, the SCAGE and ntv-Src group is the SCAGE biosensor group together with the kinase-dead c-Src (ntv-Src), and the SCAGE w/PP1 Pretreat group is the SCAGE biosensor group pretreated with 10 μM PP1 for 1 h (n = 53, 18, 28, 30, 17, 13, 10, 12, 52, 15, 17, 12, 36, 19, and 113 cells for Ori, CD, CA, GE, CD/GE, CR/GE, CT/GE, CV/GE, SCAGE, CA/GE/TV, CA/GE/KL, CA/GE/TV/KL, CA/GE/YF, SCAGE and ntv-Src and SCAGE w/PP1 Pretreat groups, respectively). (d) ECFP/FRET emission ratios of the SCAGE biosensor in Src/Yes/Fyn triple-knockout (SYF −/−) mouse embryonic fibroblasts (MEFs) cotransfected with various Src family kinases and mutants. SYF −/− MEFs were reconstituted with active-Src (n = 46), wt-Src (n = 44), ntv-Src (n = 24), wt-Fyn (n = 90), wt-Yes (n = 107), and an empty vector (n = 81). (c, d) In scatter plots, the red lines indicate the mean values, the error bars represent SEM and average data are presented as mean ± SEM; ***P < 0.001, **P < 0.01, and “n.s.”P > 0.05 are from Kruskal-Wallis test followed by Dunn’s multiple comparison test.