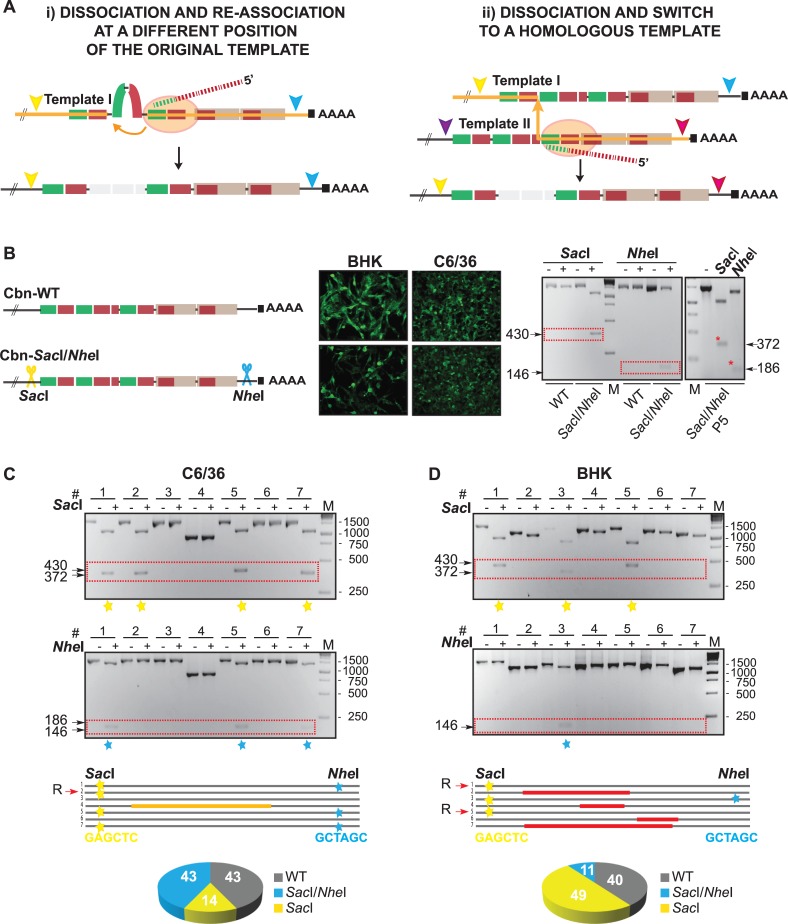
Fig 4. RNA recombination by switching of viral polymerase between homologous template strands.
(A) Schematic representation of copy-choice recombination mechanisms, where the viral RNA-dependent RNA-polymerase (orange oval) and the nascent strand (dashed lines) jump at a different position of the original template (i) or between two homologous templates (ii). Orange lines indicate the possible route of viral polymerase. (B) To mark the CHIKV-Cbn RNA, SacI and NheI restriction sites were introduced at the 5’ and 3’ ends of the viral 3’UTR, respectively. Immunofluorescence of Cbn-WT and Cbn-SacI/NheI viruses were performed in BHK and C6/36 cells. Viral RNA was recovered from the supernatant, and fragments corresponding to the last ~1,500 nucleotides of the viral genome were obtained by RT-PCR, digested with SacI and NheI restriction enzymes and analyzed by agarose gel electrophoresis. Undigested (-) and digested (+) products are shown on the right panel for parental (WT) and marked (SacI/NheI) viruses. Also, we show the digested products for one representative individual clone of SacI/NheI virus containing full-length 3’UTR, which was isolated after 5 passages in C6/36 cells (red asterisks). For P5, 20 clones were tested and they all conserved both SacI and NheI restriction sites. (C and D) Cbn-WT and Cbn-SacI/NheI RNAs were co-transfected at a 1:1 ratio into C6/36 and BHK cells, in duplicates. At 72hpt, supernatants were harvested, subjected to RT-PCR and blunt-end cloned. For each experiment, 14 individual clones were subjected to PCR reactions, enzymatic digestion, and resolved by agarose gel electrophoresis. To illustrate, undigested and digested products from 7 representative clones are shown. Depending on the orientation of each inserted fragment into the blunt plasmid, digestion of SacI positive clones generated 430 or 372 nucleotide-length products and digestion of NheI positive clones generated 186 or 146 nucleotide-length products. Presence of SacI and/or NheI restriction sites are indicated with yellow and light blue stars, respectively. Individual clones are also schematized and deletions are indicated with orange and red lines. Recombinant clones generated by template switching between strands are indicated with a R. Relative abundance of each virus is represented in pie charts (bottom). Unmarked WT 3'UTRs are indicated in grey, marked 3'UTRs sensitive to SacI and NheI digestion in light blue, and 3'UTRs that are only sensitive to SacI digestion in yellow.