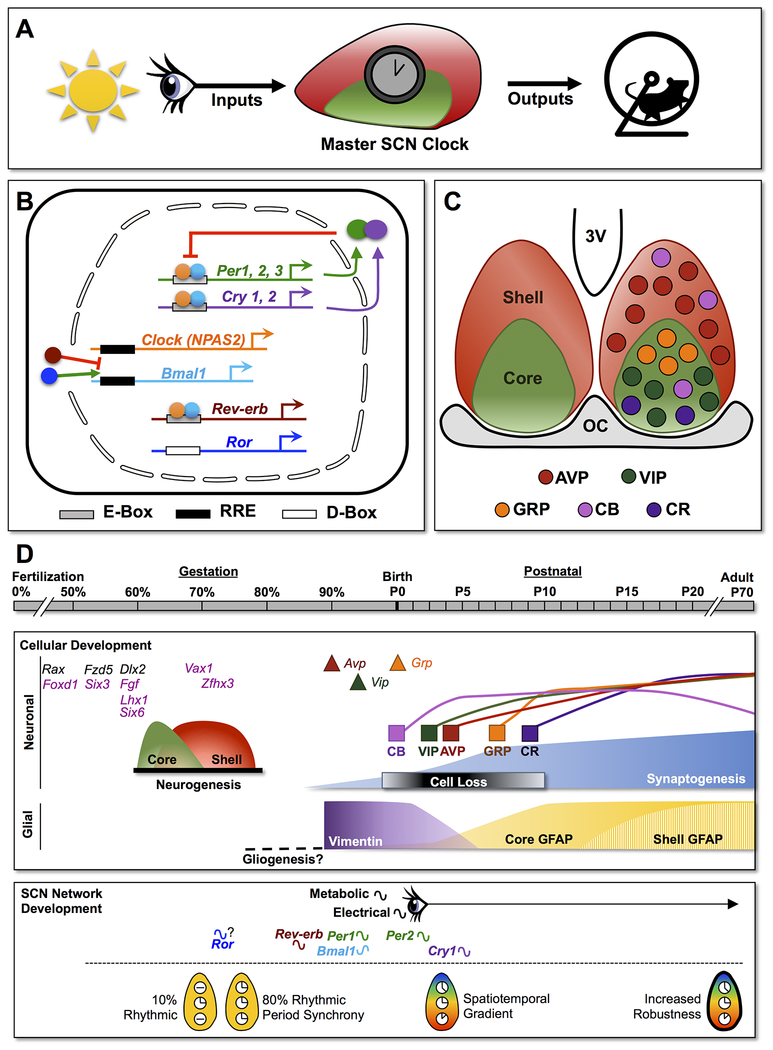
Figure1.
Development of the circadian timekeeping system. A. In adulthood, the suprachiasmatic nucleus (SCN) receives light input via the retinohypothalamic tract and provides daily outputs signals to downstream tissues to coordinate the timing of overt rhythms. B. Simplified model of the circadian molecular clock in mammals, which is composed of self-sustained transcriptional-translational feedback loops that regulate daily expression of clock genes and their protein products. In SCN neurons, the transcription factors CLOCK and BMAL1 dimerize and activate Period (Per) and Cryptochrome (Cry) expression during the day. After translation, PER and CRY dimers repress their own transcription at night. This core feedback loop is interlocked with other transcriptional loops that stabilize and augment circadian function. For example, REV-ERB and ROR regulate the daily expression of Bmal1. C. The shell-core model of the adult SCN network illustrating the spatial location of five neuronal subclasses in mice. AVP: Arginine Vasopressin, VIP: Vasoactive Intestinal Polypeptide, GRP: Gastrin-Releasing Peptide, CB: Calbindin; CR: Calretinin. Note: all five peptides are also expressed in rats and hamsters although the expression of CB and CR differs among rodent species. D. Timeline of SCN development. Important milestones are illustrated for SCN development in rodents. Most information illustrated on the timeline derives from studies using mice, but detailed information on the timing of fetal metabolic/electrical rhythms, Rev-erb rhythms, synaptogenesis, and glial maturation are only available for rats. Early genetic markers of SCN differentiation are labeled purple to represent those for which effects on SCN development have been reported. Color version of this figure can be viewed online.