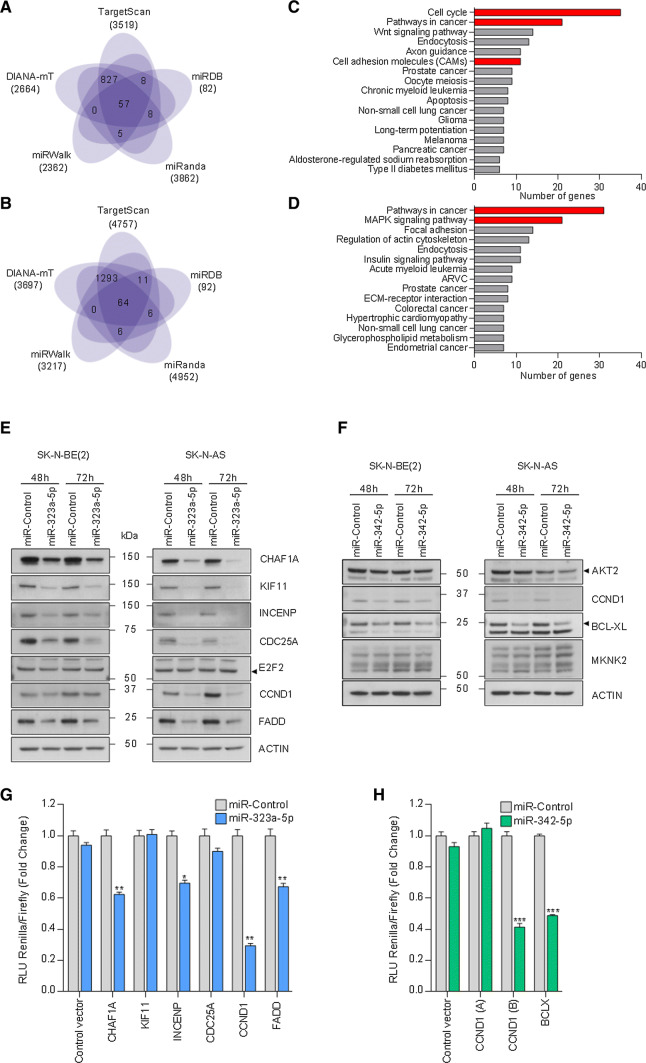
Fig. 5.
MiR-323a-5p and miR-342-5p modulated the expression of multiple cancer-related genes. a, b Venn diagram representing the overlap of predicted target genes among five miRNA-target prediction algorithms of miR-323a-5p or miR-342-5p. c, d Representation of the functional annotation of predicted target genes of miR-323a-5p (c) or miR-342-5p (d) using KEGG pathways and Gene Ontology databases. Red bars indicate selected pathways to analyze potential miRNA targets. e, f Representative Western blot of predicted target genes in SK-N-BE(2) and SK-N-AS transfected with miR-control, miR-323a-5p (e) or miR-342-5p (f) at 48 h and 72 h post-transfection. g, h Luciferase 3′UTR reporter assays. Graph represents luciferase activity in HEK-293T cells co-transfected with 50 ng/well of the indicated reporter vectors and 25 nM of miR-control, miR-323a-5p (g) or miR-342-5p (h). Data represented the average ± SEM of three independent experiments (n = 3 per experiment). *p < 0.05, **p < 0.01, ***p < 0.001