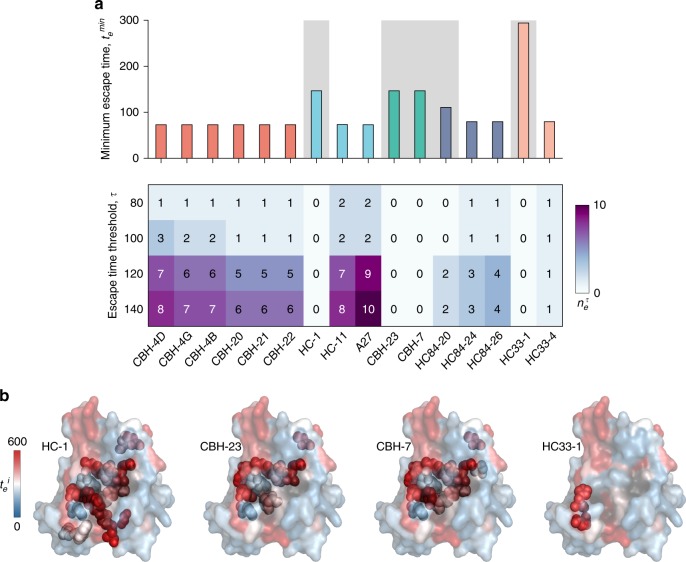
Fig. 4.
Analysis of relative escape times associated with mutating the binding residues of known HmAbs identifies potential difficult to escape HmAbs. a Minimum escape time across all residues (top panel) and number of residues with relatively short escape times (bottom panel) in the binding residues of 16 HmAbs, determined using global alanine scanning38. The binding residues of the HmAbs are defined as the mutations that resulted in reduction of binding with respect to the wild-type to ≤ 20%. HmAbs are colored according to the antigenic domains using the scheme in Fig. 3b. The background of the HmAbs predicted to be relatively escape-resistant () is shaded. b Location of binding residues of four HmAbs identified as relatively escape-resistant are shown as spheres on the available crystal structure (PDB ID 4MWF43). Heat map shows the escape times () associated with incurring mutations at antibody-binding residues similar to Fig. 3