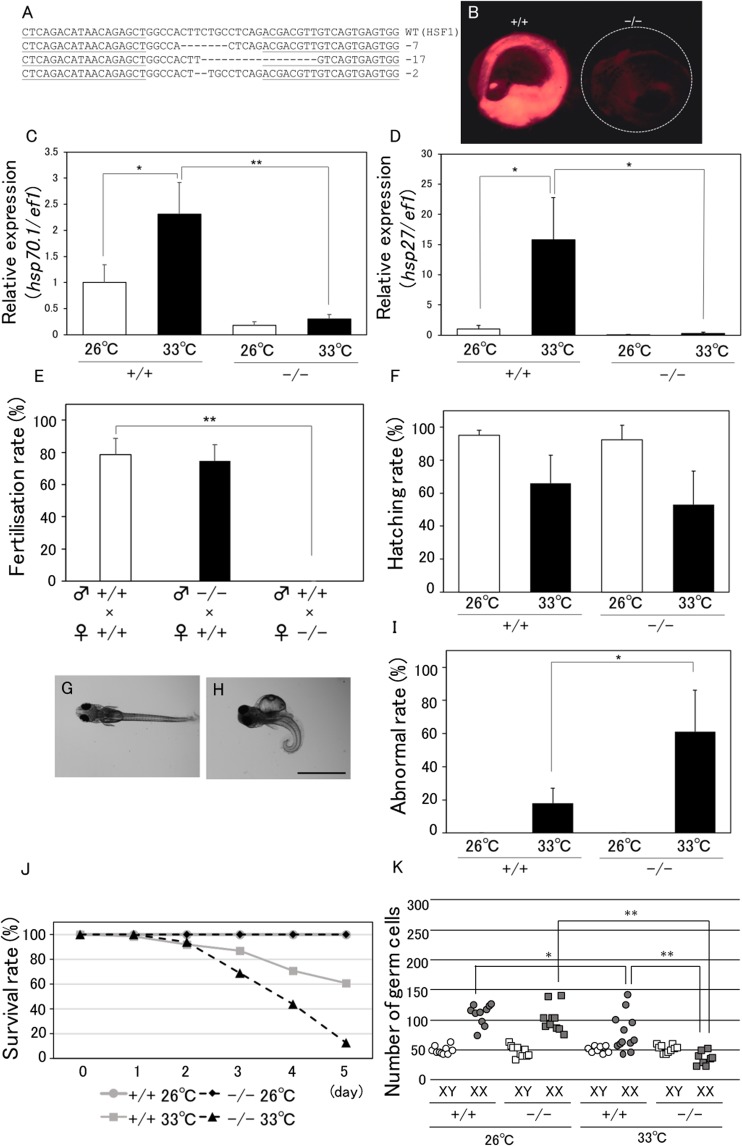
Figure 1.
Phenotypes of HSF1 KO medaka. (A) Wild-type (WT) sequences of medaka hsf1 and recognition sites of TALENs (underlined). The sizes of the deletions are shown to the right of each mutated sequence (−, deletions). (B) hsp70-DsRed Tg medaka enabling the monitoring of HSP70 expression by DsRed fluorescence. WT (+/+) and HSF1 homozygous mutated (–/–) embryos heated at 37 °C for 1 h were observed for DsRed fluorescence 1 day after heating. (C,D) hsp70 (C) and hsp27 (D) expression in whole-body fry at 0 dph. Relative expression was calculated based on the value of ef1. n = 4. (E) Fertilisation rate (fertilised eggs over obtained eggs) for 5 days in each of three mating groups: WT males and WT females, KO males and WT females, and WT males and KO females. n = 3. (F) Hatching rate (hatched fry over obtained embryos) in WT (+/+) and KO (–/–) embryos incubated at 26 °C or 33 °C. (G–I) Representative normal (G) and abnormal (H) fry incubated at 33 °C during 0 dpf to 0 dph, and the rate of abnormal fry (abnormal fry over hatched fry) (I). Scale bar, 1 mm. (J) Survival rate in WT (+/+) and KO (−/−) fry incubated at 26 °C or 33 °C during 0 dph to 5 dph. (K) Number of germ cells in WT (+/+) and KO (−/−) fry incubated at 26 °C or 33 °C during 0 dpf to 0 dph. *p < 0.05, **p < 0.01.