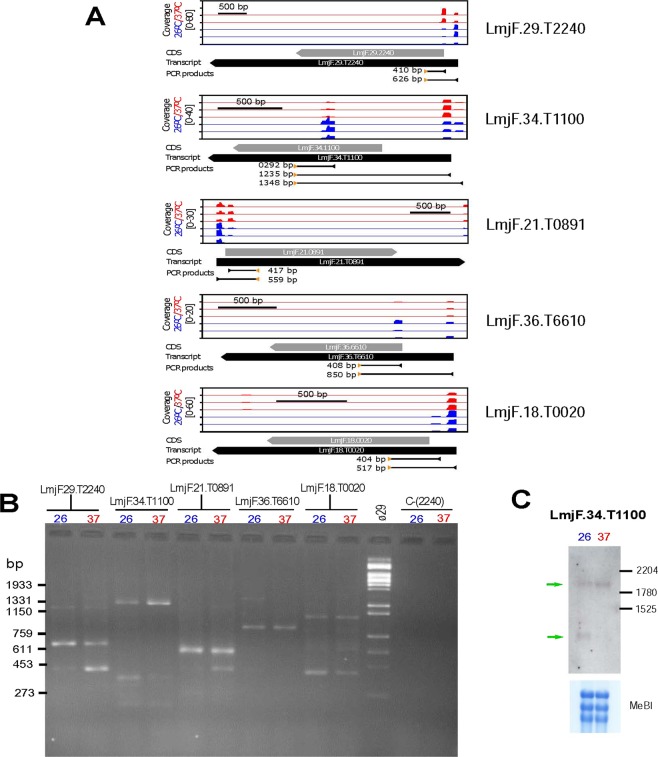
Figure 7.
Changes in the selection of SL-addition sites (SASs) associated with the temperature treatment in L. major promastigotes. (A) Schematic representation of the SASs used for trans-splicing of transcripts LmjF.29.T2240, LmjF.34.T1100, LmjF.21.T0891, LmjF.36.T6610 and LmjF.18.T0020 as determined by position of SL-containing RNA-seq reads for each one of the three experimental replicates from either promastigotes grown at 26 °C (blue) or promastigotes incubated for 2 h at 37 °C (red). For each transcript, the relative coverage is shown according to the scale indicated. Below each graph, the position of the CDS (grey arrow), transcript (black arrow) and the size and position of the expected PCR amplification products are shown. (B) PCR amplification of cDNA synthetized from RNA samples of parasites incubated at either 26 °C or 37 °C. For each transcript, a specific oligonucleotide and a common SL-oligonucleotide were used (oligonucleotide sequences are indicated in the Methods section). The negative control, C-(2240), consisted of the amplification from the RNA samples (without retrotranscription step) using the LmjF.29.T2240-specific oligonucleotide and the SL-oligonuclotide. HindIII-digested DNA of bacteriophage Φ29 was used as size marker (lane Φ29), and the size of relevant bands is indicated on the left. (C) Analysis by Northern blotting of transcripts derived from gene LmjF.28.3032 in RNA samples derived from parasites incubated at 26 or 37 °C. Bottom, methylene blue (MeBl) staining of the membrane used for hybridization. Uncropped images shown in Supplementary Figure 2.