Figure 1.
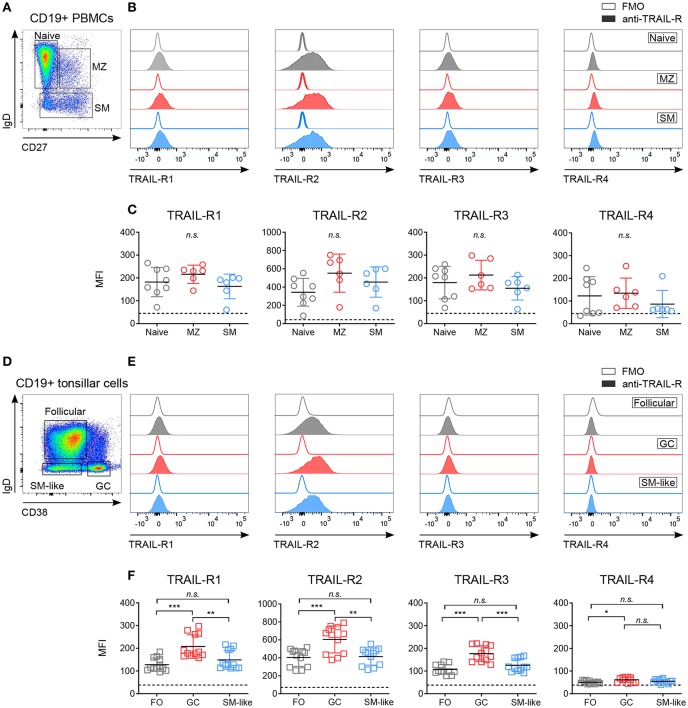
Differential expression of TRAIL-Rs in human B cell subpopulations. Expression of TRAIL-R1, TRAIL-R2, TRAIL-R3 and TRAIL-R4 was characterized by flow cytometry in ex vivo CD19+ human B cells isolated from PBMCs (A–C) and tonsillar CD19+ cells (D–F). (A) Gating strategy to identify naïve (IgD+ CD27−), marginal zone (MZ; IgD+ CD27+) and switched memory (SM; IgD− CD27−) cells from CD19+ live B cells. (B) Histograms show expression levels of indicated TRAIL-Rs in naïve, MZ and SM B cells compared to respective fluorescent minus one (FMO) controls. (C) Mean fluorescence intensity (MFI) of TRAIL-Rs among indicated B cell subsets. Each dot represents a biological replicate. (D) Gating strategy to identify follicular (FO; IgD+ CD38−), germinal center (GC; IgD− CD38+) and switched memory-like (SM-like; IgD− CD38−) cells among CD19+ live B cells. (E) TRAIL-R expression in FO, GC and SM-like B cells compared to respective FMO controls. (F) Quantifications of TRAIL-R expression among tonsillar B cell subpopulations depicted as MFI. Dots represent duplicates of six biological replicates. (C,F) Dashed lines indicate averaged FMO controls. Mean values ± SD are shown. n.s., not significant; *adj.p < 0.05; **adj.p < 0.01; ***adj.p < 0.001. Statistical analysis was performed using one-way ANOVA, followed by Tukey's multiple comparison test where appropriate.