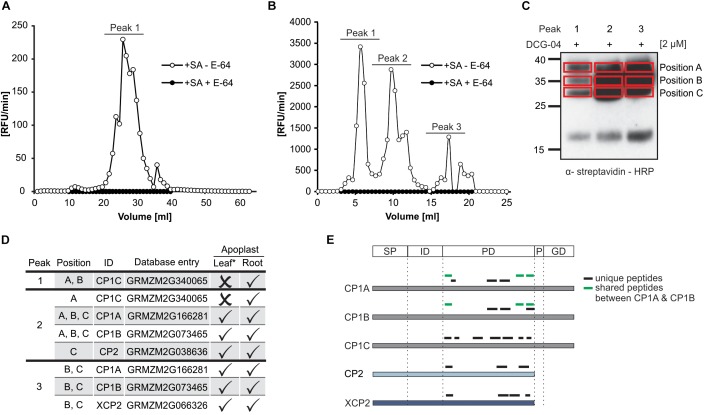
FIGURE 1.
Identification of maize root PLCPs after ion-exchange chromatography. (A) Leaf apoplastic fluid (LAF) and (B) root apoplastic fluid (RAF) of maize leaves or roots, pre-treated with 2 mM salicylic acid (+SA) were isolated and fractionated using ion exchange chromatography (IEC). Fractions were tested for PLCP activity using 10 μM of the substrate Z-FR-AMC with (+) or without (-) 2 μM E-64. The release of AMC (relative fluorescent unit = RFU) per minute was measured and plotted against fraction volume. Peaks represent fractions of higher PLCP activity that were inhibited when using E-64. (C) DCG-04 labeling of fractions corresponding to different peaks (B) were pooled and labeled with 2 μM DCG-04 for 2 h. Samples were separated by SDS-PAGE and detection of biotinylated proteins was performed using an α-streptavidin-HRP antibody. Red squares mark samples subjected to in gel digest (IGD) and MS analysis. Peaks 1, 2, and 3 correspond to pooled fractions (B) and position A, B, and C mark size separated signals of each peak loaded. (D) Identification of apoplastic root PLCPs and comparison with leaf proteome. Apoplastic root-PLCPs identified by MS analysis were compared to PLCPs previously published in leaf apoplastic fluid by van der Linde et al., 2012a (∗). Peak and position (B,C) of PLCPs identified in this study as well as PLCP presence (✓) or absence (×) is indicated. (E) Analysis of peptides found by mass spectrometry. Displayed is a schematic representation of the identified PLCPs and the position of peptides found: SP, signal peptide; ID, autoinhibitory prodomain; PD, protease C1-domain; P, proline-rich domain; GD, granulin domain. Unique peptides are labeled in black and shared peptides are labeled in green.