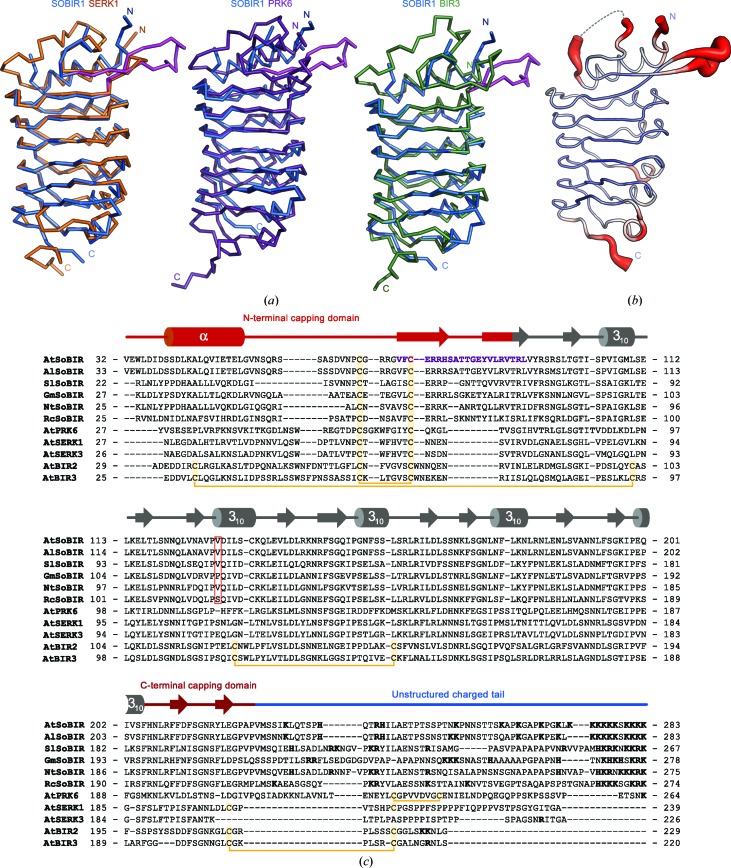
Figure 4.
AtSOBIR1 shares a common architecture with other small plant LRR-RKs. (a) Cα traces of structural superposition of the ectodomain of SOBIR1 (blue) with AtSERK1 (left, orange; PDB entry 4lsc; Santiago et al., 2013 ▸), AtPRK6 (centre, purple; PDB entry 5yah; Zhang et al., 2017 ▸) and AtBIR3 (right, green; PDB entry 6fg8; Hohmann, Nicolet et al., 2018 ▸). The SOBIR1-unique extended β-hairpin is highlighted in magenta. (b) Ribbon diagram of the SOBIR1 ectodomain with Cα atoms coloured according to their crystallographic temperature factors (red, high; blue, low). Residues 57–63, which are missing in the structure, are indicated by a dotted line. The N- and C-termini as well as an N-terminal capping-domain loop and the extending β-hairpin appear to be flexible, in contrast to the rigid and well ordered LRR core. (c) Structure-based sequence alignment of the ectodomains of SOBIR1 from A. thaliana [UniProt (http://www.uniprot.org) identifier Q9SKB2], A. lyrata (UniProt identifier D7LEA5), Solanum lycopersicum (UniProt identifier K4C8Q3), Glycine max (UniProt identifier I1JXE0), Nicotiana tabacum (UniProt identifier Q8LP72) and Ricinus communis (UniProt identifier B9RAQ8) as well as A. thaliana SERK1 (UniProt identifier Q94AG2), SERK3 (UniProt identifier Q94F62), BIR2 (UniProt identifier Q9LSI9) and BIR3 (UniProt identifier O04567). Shown alongside is a secondary-structure assignment (calculated with DSSP; Kabsch & Sander, 1983 ▸), with the N- and C-terminal capping domains highlighted in red and a unstructured region at the C-terminus shown in blue. Disulfide bridges are shown in yellow and the SOBIR1-specific β-hairpin in purple; the position of Val129 of AtSOBIR1, which is mutated to methionine in sobir1-8, is indicated by a red box; positively charged residues in the unstructured C-terminal tail are highlighted in bold.