Abstract
Chemical probes of epigenetic ‘readers’ of histone posttranslational modifications (PTMs) have become powerful tools for mechanistic and functional studies of their target proteins in normal physiology and disease pathogenesis. Here we report the development of the first class of chemical probes of YEATS domains, newly identified ‘readers’ of histone lysine acetylation (Kac) and crotonylation (Kcr). Guided by the structural analysis of a YEATS-Kcr complex, we developed a series of peptide-based inhibitors of YEATS domains by targeting a unique π-π>-π >stacking interaction at the proteins’ Kcr recognition site. Further structure optimization resulted in the selective inhibitors preferentially binding to individual YEATS-containing proteins including AF9 and ENL with submicromolar affinities. We demonstrate that one of the ENL YEATS-selective inhibitors, XL-13m, engages with endogenous ENL, perturbs the recruitment of ENL onto chromatin, and synergizes the BET and DOT1L inhibition-induced down-regulation of oncogenes in MLL-rearranged acute leukemia.
Posttranslational modifications (PTMs) of histones play essential roles in regulating gene functions1. The addition and removal of most histone PTMs are dynamically controlled by histone-modifying enzymes (known as ‘writers’ and ‘erasers’)2. Histone PTMs (or marks) can serve as the docking sites for effector proteins (‘readers’) that specifically recognize them3–5. By recruiting ‘readers’ to the chromatin, histone PTMs signal the downstream biological events (e.g., gene transcription, DNA replication and repair), which underlies one of the key cellular mechanisms for epigenetic regulation6,7. Given their importance in maintaining normal cell physiology, improper ‘writing’, ‘erasing’ and ‘reading’ of histone modifications have been implicated in many human diseases, such as cancer8,9. The epigenetic protein families have therefore emerged as promising targets for drug discovery10,11. The development of chemical epigenetic modulators, mostly inhibitors, has provided useful tools to probe the regulatory mechanisms and biological significances of histone PTMs. More importantly, a collection of inhibitors has become potential therapeutic agents for the treatment of diseases associated with abnormal epigenetic regulation.
Early endeavors targeting the epigenome have been focusing on histone-modifying enzymes, leading to the development of potent and selective inhibitors12. Several histone deacetylase (HDAC) inhibitors have even been approved for clinical uses (e.g., vorinostat)13. The inhibitor development for epigenetic ‘readers’ has been a relatively slow-paced process. One possible reason could be that the recognition patterns of histone PTMs by the readers commonly involve shallow binding pockets or even appear as surface groove recognitions. The affinity for such PTM-mediated interactions are normally weak, and the binding pockets are seldom ideal for inhibitors with nanomolar activities. In addition, the design of inhibitors targeting the interfaces with a large area is much more challenging than targeting an enzyme with well-defined catalytic residues. Bromodomain (BrD), which recognizes lysine acetylation (Kac, Supplementary Fig. 1a), is one of the epigenetic ‘readers’ that has been extensively explored for inhibitor development14. A set of BrD inhibitors are now in different stages of clinical trials for the treatment of human diseases, including cancer, atherosclerosis, as well as diabetes15,16.
Besides the BrDs, YEATS domain was identified as a novel ‘reader’ of Kac17,18. The human genome encodes 4 YEATS domain-containing proteins, which are AF9, ENL, YEATS2, and GAS4119. Unlike the BrDs with end-closed binding cavities, the YEATS domains coordinate the acetyllysine with an end-open aromatic ‘sandwich’ cage shaped by two conserved aromatic residues17. Notably, the tunnel-like feature of the end-open cage also enables the YEATS domains to recognize the histone lysine crotonylation (Kcr, Supplementary Fig. 1a) marks20–23, whose acyl chain is two-carbon longer than Kac. Moreover, the conjugated O=C−C=C moiety of the crotonyl group forms a unique π-π-π stacking with the two conserved aromatic residues, which significantly enhances the binding affinity, suggesting a preference of YEATS domain toward Kcr over Kac. Recently, AF9 YEATS domain was found to mediate active transcription through ‘reading’ the histone Kac and Kcr marks17,20. The YEATS-dependent association of ENL with acetylated histone H3 was reported to be essential for oncogenic gene expression in aggressive leukemia24,25. By ‘reading’ the H3K27ac mark, YEATS2 regulates transcriptional programs essential for tumorigenesis of non-small cell lung cancer26. Despite these discoveries, we still lack comprehensive understanding of the functional outcomes of YEATS-Kac/Kcr interactions in the epigenetic regulation. The development of YEATS domain inhibitors should provide useful tools to fill such knowledge gap, and offer potential therapeutic agents targeting the YEATS domain.
Here we describe the structure-guided development of, to the best of our knowledge, the first class of YEATS domain inhibitors by targeting the π-π-π stacking in the aromatic ‘sandwich’ cage. Structure optimization led to the identification of AF9 YEATS- and ENL YEATS-selective inhibitors with potent submicromolar activity. We showed that an ENL YEATS-selective inhibitor, XL-13m (1), engaged with endogenous ENL. Using MOLM-13 cells, an MLL-rearranged acute leukemia cell line, we also demonstrated that XL-13m can be used as a chemical probe to interrogate the YEATS-dependent role of ENL in regulating gene transcription.
RESULTS
Targeting YEATS domain by enhanced π-π-π stacking
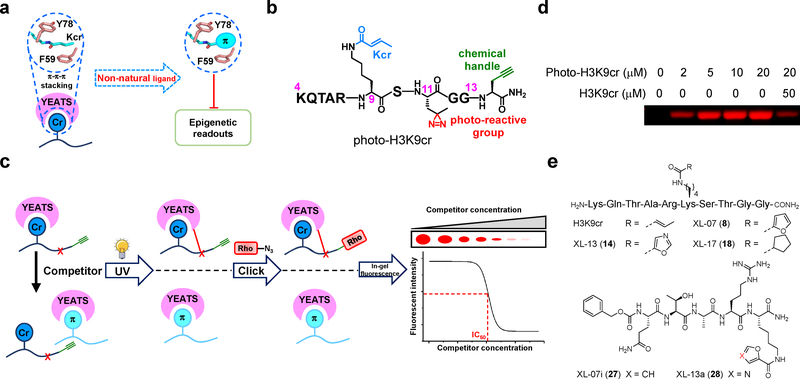
Our design of YEATS domains inhibitors was enlightened by the unique π-π-π stacking in the crystal structure of AF9 YEATS-H3K9cr complex20. In this structure, the conjugated crotonyl group inserts into the aromatic ‘sandwich’ cage formed by the two conserved aromatic residues, F59 and Y78 (Fig. 1a and Supplementary Fig. 1b). This unique π-π-π interaction underlies why the binding affinity of YEATS domains to Kcr is normally 2−5 folds higher than to Kac27. We reasoned that replacing the crotonyl group with an expanded π system may further enhance the interaction. Therefore, to target the YEATS domains, we designed and synthesized a series of decapeptides XL-01 to XL-16 (2 to 17, Supplementary Fig. 1c) derived from histone H3 (residues 4–13) with Lys 9 carrying π system-containing functional groups.
Figure 1. Targeting the π-π-π stacking for YEATS inhibition.
(a) Design strategy of peptide-based YEATS domain inhibitors. Introducing expanded π system-containing functional groups to replace crotonyl side chain for YEATS inhibition. (b) Chemical structure of photoaffinity probe photo-H3K9cr. (c) Schematic diagram illustrating the competitive photo-cross-linking assay for determination of inhibitory effects of developed oligopeptides against YEATS domains. (d) Recombinant AF9 YEATS was robustly captured by photo-H3K9cr, and the labeling was competed off by the H3K9cr peptide. Photo-cross-linking experiment was repeated independently for two times with similar results. (e) Chemical structures of representative oligopeptides developed for targeting YEATS domain. Uncropped gels are provided in Supplementary Fig 12.
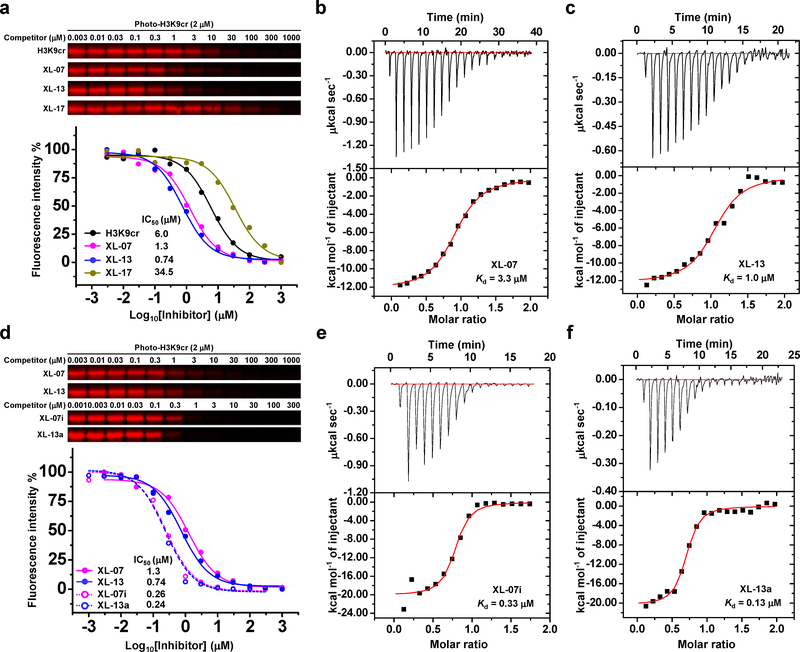
We evaluated the binding ability of the synthesized decapeptides toward the YEATS domains using a competitive photo-cross-linking assay28–30. The assay relied on a photoaffinity probe (photo-H3K9cr, Fig. 1b) that covalently captures the YEATS domains upon UV irradiation. Photo-H3K9cr was derived from a H34-13K9cr peptide, in which the Thr 11 was replaced by a diazirine-containing photoreactive amino acid (photo-Leu). The probe also had a terminal alkyne-containing amino acid (propargylglycine) to enable bioorthogonal conjugation of fluorescence tags for detection of the captured proteins. If a tested decapeptide could compete with photo-H3K9cr to occupy the same Kcr-binding sites of the YEATS domains, the labeling of YEATS domains would be inhibited (Fig. 1c). As shown in Fig. 1d, photo-H3K9cr robustly labeled AF9 YEATS in a concentration-dependent manner. This labeling was competed off by the native H3K9cr peptide with an IC50 of 6.0 μM (Supplementary Fig. 2a), which is in a line with its dissociation constant (Kd = 14.7 μM, Supplementary Fig. 3a and 3b) determined by isothermal titration calorimetry (ITC). We then tested the decapeptides XL-01 to XL-16. Strikingly, 6 out of the 16 decapeptides showed lower IC50 than the native H3K9cr peptide toward the AF9 YEATS (Supplementary Fig. 2b–2q). The two decapeptides carrying a 2-furancarbonyl side chain (XL-07, Fig. 1e) and a 5-oxazolecarbonyl side chain (XL-13, Fig. 1e) showed the highest inhibition potencies with IC50 values of 1.3 and 0.74 μM, respectively (Fig. 2a, Supplementary Fig. 2h and 2n). In contrast, a control decapeptide XL-17 (18, Fig. 1e) lacking a conjugated π system displayed a very weak inhibitory effect (Fig. 2a and Supplementary Fig. 2r), highlighting the crucial role of the expanded π system for targeting the YEATS domains. Further ITC measurements showed that the replacement of the crotonyl group (Kd = 14.7 μM) by 2-furancarbonyl (XL-07, Kd = 3.3 μM, Fig. 2b and Supplementary Fig. 3b) and 5-oxazolecarbonyl (XL-13, Kd = 1.0 μM, Fig. 2c and Supplementary Fig. 3b) led to 4.5- and 14.7-fold binding enhancement, respectively.
Figure 2. Characterization and optimization of inhibitory effects.
(a) In-gel fluorescence and competition curves of representative decapeptides H3K9cr, XL-07, XL-13, and XL-17 against AF9 YEATS domain obtained from competitive photo-cross-linking assay. (b-c) ITC measurements for the binding affinities of AF9 YEATS toward (b) XL-07 and (c) XL-13, respectively. (d) Comparison of the inhibitory effects of XL-07i and XL-13a and their parent decapeptides against AF9 YEATS by competitive photo-cross-linking assay. (e-f) ITC measurements for the binding affinities of AF9 YEATS toward (e) XL-07i and (f) XL-13a, respectively. For all the competitive photo-cross-linking assay, after UV irradiation, the photo-H3K9cr-labeled protein was conjugated to rhodamine-N3 and visualized by in-gel fluorescence scanning. The fluorescence intensity of each band was quantified by ImageJ. All curves were normalized between 100% and 0% at the highest and lowest fluorescence intensities, respectively. Data are reported as mean of two independent experiments. All the ITC measurements were repeated independently for three times with similar results. Thermodynamic parameters were listed in Supplementary Fig. 3b. Uncropped gels are provided in Supplementary Fig 12.
Toward minimal length and higher potency
We next sought out to optimize the developed inhibitors toward a shorter length and higher potency. In the AF9 YEATS-H3K9cr complex, the H3 Arg 8 and Lys 9 residues provide the key contacts and dominate the interaction with the YEATS domain (Supplementary Fig. 1b)20. We therefore synthesized a dipeptide XL-07a (19) with the RK signature motif (Supplementary Fig. 4a) and added to it the surrounding residues one by one to generate another 7 inhibitors XL-07b to XL-07h (20 to 26, Supplementary Fig. 4a). Comparing with XL-07, the dipeptide XL-07a showed a significantly reduced inhibitory activity (Supplementary Fig. 4b and 4c). Adding the residues at the N-terminus of XL-07a gradually restored the inhibitors’ potency. The pentapeptide XL-07h with the QTARK sequence exhibited a comparable activity (IC50 = 4.5 μM, Supplementary Fig. 4d) with XL-07. In contrast, the C-terminal residues of the RK motif negatively impacted their interactions with AF9 YEATS (Supplementary Fig. 4b and 4c). Capping the N-terminus of the pentapeptide XL-07h with a hydrophobic carboxybenzyl (Cbz) group produced a more potent inhibitor XL-07i (27, IC50 = 0.26 μM, Kd = 0.33 μM, Fig. 1e, 2d and 2e, Supplementary Fig. 3b and 4e). Replacing the 2-furancarbonyl group in XL-07i with 5-oxazolecarbonyl group generated another inhibitor XL-13a (28, Fig. 1e) with slightly strengthened potency (IC50 = 0.24 μM, Kd = 0.13 μM, Fig. 2d and 2f, Supplementary Fig. 3b and 4f).
Crystallographic studies of AF9 YEATS-inhibitor complex
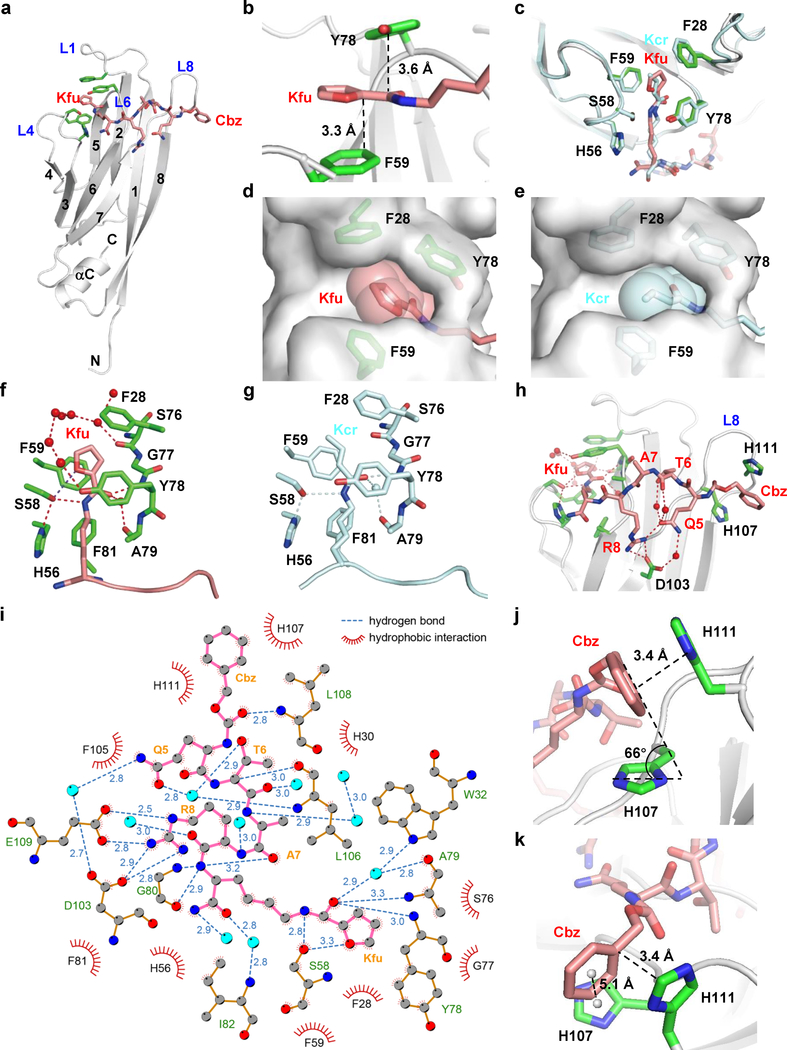
To study the molecular basis underlying AF9 YEATS inhibition, we determined the crystal structure of AF9 YEATS bound to XL-07i at 1.9 Å resolution (Supplementary Table 1). In the complex, XL-07i stretches along the surface formed by loops L4, L6, and L8 of AF9 YEATS (Fig. 3a). The planar 2-furancarboyl group inserts into the same Kac/Kcr-binding aromatic ‘sandwich’ cage and parallels well with the aromatic rings of AF9 F59 and Y78, forming an expected π-π-π stacking (Fig. 3b). Comparing with the Kcr accommodation, the aromatic cage is almost identically arranged to coordinate the 2-furancarbonyl moiety of XL-07i, except for a slight conformational change of the F28 phenyl ring (Fig. 3c). The substitution of crotonyl by 2-furancarbonyl expands the interaction surface area from 174 Å2 to 235 Å2, filling the unoccupied space in the binding pocket, and ensuring a snug encapsulation of the 5-membered furan ring (Fig. 3d and 3e). Besides retaining the conserved hydrogen bonding interactions in crotonylamide recognition, the furan oxygen atom forms one more hydrogen bond with the side chain hydroxyl group of AF9 S58, further stabilizing the 2-furancarbonyl group (Fig. 3f and 3g). The residues other than the modified lysine in XL-07i form extensive polar and hydrophobic contacts with AF9 YEATS (Fig. 3h and 3i), strengthening the protein-inhibitor interaction. Notably, the geometry between the XL-07i Cbz group and the H107 and H111 residues in loop L8 of AF9 YEATS suggests that the phenyl ring of Cbz forms parallel-displaced (with H111) and edge-to-face (with H107) π stackings with the two imidazole rings, respectively (Fig. 3j and 3k). These stackings are likely to be the reason that the addition of the N-terminal Cbz group largely increased the inhibitory potency of the inhibitors.
Figure 3. Molecular basis underlying AF9 YEATS and XL-07i interaction.
(a) Overall structure of AF9 YEATS bound to XL-07i in ribbon view. (b) The π system of XL-07i forms the expected π-π-π stacking with F59 and Y78 of AF9. (c) Superimposition of the aromatic ‘sandwich’ cage in XL-07i-bound AF9 YEATS with H3K9cr-bound AF9 YEATS. (d-e) Comparison of cavity encapsulation of (d) Kfu and (e) Kcr shown in surface view. (f-g) Hydrogen bonding networks in the binding pockets with (f) Kfu and (g) Kcr. (h) Detailed hydrogen-bonding interaction network between AF9 YEATS and XL-07i. (i) LIGPLOT diagram illustrating the contacts between XL-07i and the AF9 YEATS domain. XL-07i (pink) and key residues of AF9 YEATS (gold) are depicted in ball-and-stick mode. Grey ball, carbon; blue ball, nitrogen; red ball, oxygen; cyan ball, water molecule. Numbers showing the distance of indicated hydrogen bonds in Å. (j-k) The Cbz of XL-07i forms parallel-displaced and edge-to-face π stacking with H107 and H111 of AF9 YEATS in loop L8, respectively. For AF9 YEATS-XL-07i complex structure, AF9 YEATS (grey) is shown as ribbons, key pocket residues (green) and the XL-07i (salmon) are depicted as sticks; hydrogen bonds are shown as red dashes; the water molecules are represented as red balls; the centroids of aromatic rings are represented as grey balls. For AF9 YEATS-H3K9cr complex structure, AF9 YEATS, hydrogen bonds, and water molecules are colored and labeled cyan. Kfu: 2-furancarbonyl lysine.
In silico modeling study on AF9 YEATS-XL-13a interaction revealed that the replacement of furan ring to oxazole ring contributes additional contacts between the inhibitor and the protein. Facilitated by the nitrogen atom in the oxazole ring, XL-13a involves in the hydrogen bonding network between G77 and Y78 of AF9 and a cluster of structural water molecules nearby (Supplementary Fig. 5a). Additionally, the conformation of the 5-oxazolecarbonyl group points the nitrogen atom to F28 of AF9 in a manner that suggests a moderate lone pair-π interaction (Supplementary Fig. 5b). The water-mediated hydrogen bonding and the lone pair-π interaction make the 5-oxazolecarbonyl group more favorable to the AF9 YEATS than 2-furancarbonyl group, leading to an enhanced inhibitory activity.
Developing ENL YEATS-selective inhibitors
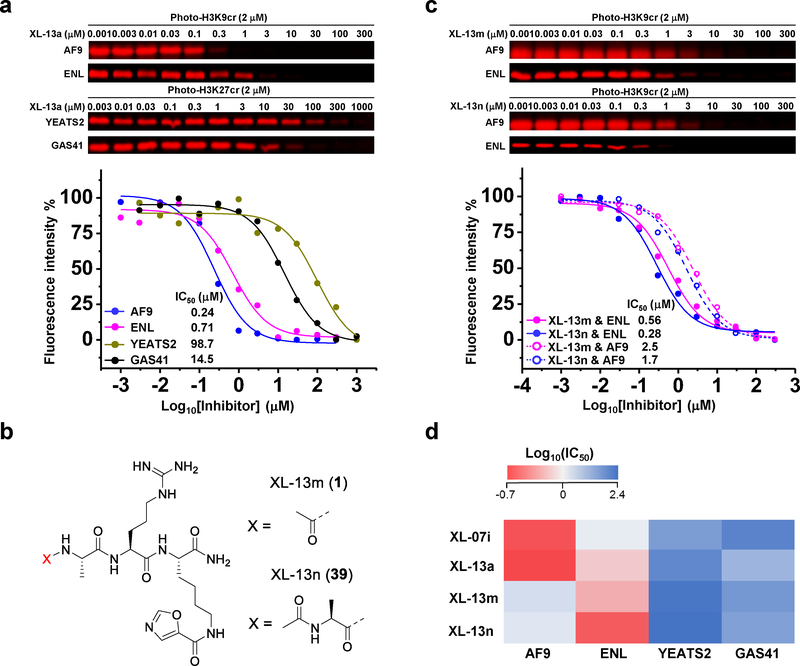
We next asked if the developed inhibitors could target the other YEATS domains. The competition assays against YEATS domains of ENL, YEATS2, and GAS41 showed that XL-07i and XL-13a were more selective toward the YEATS domains of AF9 and ENL over YEATS2 and GAS41 (Fig. 4a, Supplementary Fig. 6a–6h). Given the high sequence and structure similarity between AF9 and ENL YEATS domains, it was expected that the AF9 YEATS-binding inhibitors may also interact with ENL YEATS. However, it was interesting to learn that, comparing with AF9 YEATS, XL-07i and XL-13a exhibited 5- and 2.9-fold lower potency against ENL YEATS, respectively. This observation demonstrated the possibility to develop inhibitors that distinguish these two similar YEATS domains. It was recently reported that ENL, but not AF9, participated in the regulation of oncogenic gene transcription in acute leukemia through ‘reading’ the histone Kac marks by its YEATS domain24,25. We therefore sought to develop inhibitors that selectively target the ENL YEATS.
Figure 4. Development of ENL YEATS-selective inhibitors.
(a) Determination of XL-13a inhibitory effects on different YEATS domains by competitive photo-cross-linking. (b) Chemical structures of XL-13m and XL-13n. (c) Comparison of the inhibitory effects of XL-13m and XL-13n against AF9 and ENL YEATS domains. (d) Heat map showing that the developed YEATS inhibitors were selective for YEATS domains of AF9 and ENL over YEATS2 and GAS41. For all the competitive photo-cross-linking assay, the photo-H3K9/27cr-labeled proteins were conjugated to rhodamine-N3 and visualized by in-gel fluorescence scanning. The fluorescence intensity of each band was quantified by ImageJ. All curves were normalized between 100% and 0% at the highest and lowest fluorescence intensities, respectively. Data are reported as mean of two independent experiments. For the chemical structure of photo-H3K27cr, see Supplementary Fig. 6a. Uncropped gels are provided in Supplementary Fig 12.
The ENL YEATS domain was found to have a slightly higher affinity to bind H3K27cr than H3K9cr20. We thus prepared peptides based on the amino acids surrounding the K27 site and with the 5-oxazolecarbonyl π system installed at this lysine residue. By varying the length of the flanking sequence, another 14 oligopeptides XL-13b to XL-13o (29 to 41, Supplementary Fig. 7a) containing the signature RK motif were synthesized and screened for their inhibitory activities against ENL YEATS (Supplementary Fig. 7b and 7c). Interestingly, unlike the observation in AF9 YEATS inhibition, the dipeptide XL-13l (IC50 = 3.3 μM, Supplementary Fig. 7d) with only the RK signature motif showed even higher inhibitory efficiency than the undecapeptide XL-13b (IC50 = 6.9 μM, Supplementary Fig. 7e), indicating the dominant contribution of these two key residues in the ENL YEATS recognition. Further addition of the N-terminal residues achieved improved inhibition of ENL YEATS for the tripeptide XL-13m (IC50 = 0.56 μM) and tetrapeptide XL-13n (IC50 = 0.28 μM) (Fig. 4b and 4c, Supplementary Fig. 8a and 8b). In contrast to the AF9 YEATS-selective inhibitors XL-07i and XL-13a, these two new inhibitors showed preference to target ENL YEATS as their inhibition against AF9 YEATS were 4.5- and 6.1-fold weaker (for XL-13m, IC50 = 2.5 μM, for XL-13n, IC50 = 1.7 μM, Fig. 4c, Supplementary Fig. 8c and 8d). Furthermore, we also examined the developed YEATS inhibitors against other two human YEATS domains (YEATS2 and GAS14), Kac ‘readers’ (BrDs of CBP, BAZ2B, and BRD4), Kac/Kcr ‘eraser’ (Sirt3), and ‘readers’ of lysine methylation marks (SPIN1 and ING2). The data suggest that these inhibitors are selective to AF9 and ENL but have little inhibitory effects toward other tested epigenetic regulators. (Fig. 4d, Supplementary Fig. 8e–8h, 9a and 9b).
Engagement of XL-13m with endogenous ENL
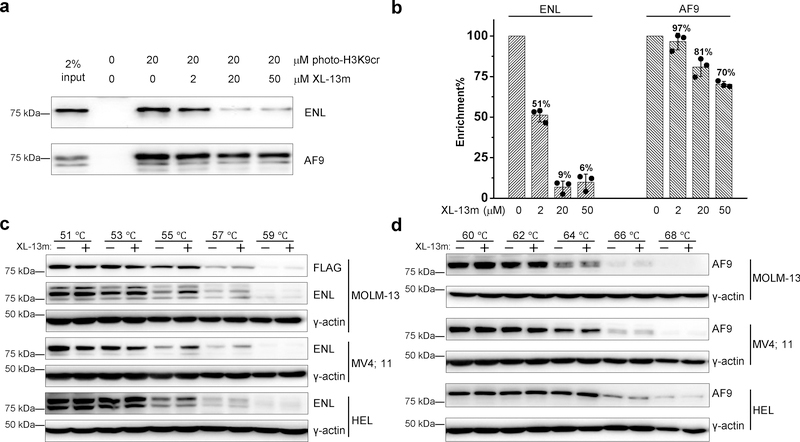
We next examined the ability of the developed inhibitors to target endogenous ENL in cells. Given the comparable potency of the tripeptide XL-13m and tetrapeptide XL-13n, we chose to further study XL-13m, which was expected to have better cell permeability. To this end, we carried out a competitive pull-down experiment, in which the cell nuclear extracts was photo-cross-linked by photo-H3K9cr in the presence or absence of XL-13m. The cross-linked proteins were then conjugated with biotin azide (biotin-N3) via click chemistry, followed by streptavidin enrichment, SDS-PAGE, and immunoblotting analysis. As shown in Fig. 5a, photo-H3K9cr successfully cross-linked and enriched endogenous AF9 and ENL proteins from the nuclear extracts of HEK 293T cells. XL-13m, at the concentration as low as 2 μM, inhibited around 50% of the photo-H3K9cr-induced enrichment of endogenous ENL. In contrast, the enrichment of AF9 was not significantly affected (~3% inhibition). At the high concentrations of 20 or 50 μM, XL-13m caused more than 90% inhibition on the ENL enrichment but less than 30% on the AF9 (Fig. 5b). This result suggests that while the AF9 YEATS-Kcr interaction could be slightly affected by XL-13m, the ENL YEATS-Kcr interaction was much more sensitive to XL-13m treatment. The competitive pull-down assays were also performed using nuclear extracts of another two cell lines, HeLa S3 and MLL-rearranged leukemia cells MOLM-13 that stably express Flag-tagged ENL at a level equivalent to the endogenous ENL protein24 (Supplementary Fig. 10). As expected, the photo-H3K9cr-induced enrichment of ENL was significantly inhibited by XL-13m, but with little effects on the enrichment of AF9 (Supplementary Fig. 11). These results agree well with the observed selectivity of XL-13m toward ENL using the recombinant YEATS domains.
Figure 5. Selective engagement of XL-13m with endogenous ENL.
(a) In-vitro photo-cross-linking pull-down in nuclear extracts (1 mg/mL) with photo-H3K9cr (20 μM) in the presence of increasing concentrations of XL-13m. Samples without adding photo-H3K9cr was prepared as negative control. After photo-cross-linking, the photo-H3K9cr-labled proteins were conjugated to biotin-N3 and enriched by streptavidin. The eluted protein mixtures were analyzed by immunoblotting against indicated antibodies. The blotting is representative of three independent experiments. (b) Quantification of the inhibitory effects of XL-13m on the enrichment of ENL and AF9 in panel a. Data are reported as mean ± s.d. (n = 3). (c-d) Cellular thermal shift assays were performed with MOLM-13 (Flag-ENL), MV4;11, and HEL cells treated with or without 50 μM XL-13m at indicated temperatures. The level changes of the soluble portion of (c) ENL and (d) AF9 were detected against indicated antibodies. The blotting is representative of two independent experiments. Uncropped blots are provided in Supplementary Fig 12.
We next applied the cellular thermal shift assay (CETSA)31,32 to examine if XL-13m was cell permeable and could target ENL in living cells. Three leukemia cell lines, MOLM-13, MV4;11, and HEL, were treated with XL-13m, and then heated at different temperatures to denature and precipitate proteins. The inhibitor-bound ENL would have better thermal stability and thus resist the high temperature-induced precipitation. Soluble proteins were extracted by freeze-thaw cycle and subjected to immunoblotting analysis. Indeed, we detected higher abundance of soluble ENL proteins in XL-13m treated samples at higher temperature (55 °C, 57 °C, and 59 °C) comparing with the untreated ones (Fig. 5c). On the contrary, the thermal stability of endogenous AF9 showed almost no differences with or without XL-13m treatment (Fig. 5d). The CETSA results in three different cell lines clearly demonstrated that XL-13m can enter cells and engage with the endogenous ENL but not AF9 under the tested condition.
XL-13m perturbs the recruitment of ENL onto chromatin
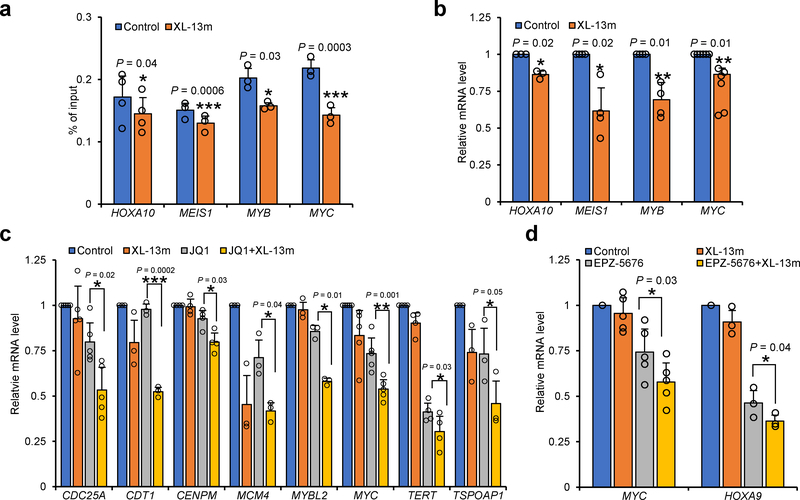
ENL was known to regulate the leukemogenic gene transcription through its YEATS domain-dependent association with chromatin24,25. We next examined whether XL-13m could perturb the ENL-chromatin interaction and thereby modulate gene transcription. By chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR), we detected the decreases in the abundance of ENL at several selected genes related to leukemia initiation, including HOXA10, MYB, MYC and MEIS1, in the MOLM-13 cells treated with XL-13m, suggesting that engagement of the inhibitor could indeed prevent the enrichment of ENL on its targeted genes (Fig. 6a). Furthermore, we observed a significant loss in the transcription levels of these pro-leukemogenesis genes upon XL-13m treatment (Fig. 6b), which is consistent with the unloading of the ENL from chromatin. Our results suggested that the XL-13m-induced YEATS inhibition blocked the chromatin association of ENL, suppressing the transcription of the oncogenes HOXA10, MYB, MYC and MEIS1 in acute leukemia cells. The observation agrees with the recent discoveries that ENL down-regulation or loss-of-Kac-binding mutation in ENL YEATS led to transcription defects of its target genes24,25.
Figure 6. XL-13m perturbs ENL-chromatin interaction and synergizes with BET and DOT1L inhibition.
(a) ChIP-qPCR analysis of ENL target genes in MOLM-13 (Flag-ENL) cells treated with or without XL-13m (50 μM) for 24 hours. Data are presented as the mean values of percentage of input ± s.e. from n = 4 (HOXA10) or n = 3 (MEIS1, MYB and MYC) independent experiments. (b) RT-qPCR analysis shows the mRNA levels of ENL target genes in MOLM-13 (Flag-ENL) cells treated with or without XL-13m (50 μM) for 24 hours. (c) RT-qPCR analysis shows the mRNA levels of selective JQ1-downregulated genes in untreated MOLM-13 (FLAG-ENL) cells, or cells treated with XL-13m (50 μM), JQ1 (50 nM) or both for 24 hours. (d) RT-qPCR analysis shows the mRNA levels of MYC and HOXA9 in untreated MOLM-13 (Flag-ENL) cells, or cells treated with XL-13m (50 μM), EPZ-5676 (500 nM) or both for 72 hours. The relative mRNA levels were normalized by that of B2M. The mean values of control groups in (b-d) were set to 1. Data are presented as mean ± s.e. from n = 3 ((b) HOXA10; (c) CDT1, MCM4, MYBL2 and TSPOAP1; (d) HOXA9) or n = 4 ((b) MEIS1 and MYB; (c) CENPM and TERT) or n = 5 ((c) CDC25A and MYC; (d) MYC) or n = 6 ((b) MYC) independent experiments. The P values are based on the two-tailed Student’s test. *P <0.05, **P <0.01, ***P <0.001.
XL-13m synergizes with BET and DOT1L inhibition
Finally, we sought to probe the regulatory roles of ENL in gene transcription using XL-13m. As a core component of the super elongation complex (SEC), ENL has been suggested to participate in transcription control together with other SEC members such as the positive transcription elongation factor (P-TEFb)24. Through direct association with P-TEFb, bromodomain and extraterminal domain (BET) family members (e.g., BRD4) serve as positive regulators of transcription33,34. A recent RNA-seq study showed that the depletion of ENL or the mutation disrupting ENL YEATS-chromatin association in leukemia cells potentiated the BET inhibitor JQ1-induced transcriptional variations24. By analyzing the reported RNA-seq data, we selected 8 candidate genes (CDC25A, CDT1, CENPM, MCM4, MYBL2, MYC, TERT, TSPOAP1) that displayed no less than 1.5-fold level changes in the JQ1-induced transcriptional down-regulation upon ENL-depletion. We then incubated MOLM-13 cells with XL-13m and JQ1, followed by real time (RT)-qPCR analysis to monitor the transcription level changes of the selected genes. The treatment of JQ1 or XL-13m, respectively, moderately lowered the transcription levels of most candidate genes (Fig. 6c). More importantly, the co-treatment of XL-13m and JQ1 significantly enhanced the transcriptional down-regulation of all 8 candidate genes caused by either XL-13m or JQ1 alone (Fig. 6c). In addition to SEC, ENL also directly interacts with histone methyltransferase DOT1L. It has been recently reported that ENL and DOT1L collaboratively regulate transcription programs responsible for the maintenance of MLL-rearranged leukemia25. In light of this discovery, we treated the MOLM-13 cells with XL-13m alone or in combination with DOT1L inhibitor EPZ-5676 and performed RT-qPCR to analyze the transcription level of MYC and HOXA9, two well-characterized genes promoting leukemogenesis. The co-treatment of XL-13m and EPZ-5676 led to an enhanced effect on the transcription suppression of MYC and HOXA9 (Fig. 6d). Our results, in mutual complement with recent studies on ENL depletion or loss-of-function mutation24,25, suggested a synergistic effect of ENL YEATS inhibition with BET and DOT1L inhibition in attenuating oncogene transcription in acute leukemia.
DISCUSSION
Chemical inhibitors of histone PTM ‘readers’ are valuable tools for elucidating regulatory mechanisms and functions of their target proteins16,35,36. While the YEATS domains were found to recognize histone Kcr marks20–23, inhibitors targeting this new class of ‘readers’ have been lacking. In this study, we developed the peptide-based inhibitors of YEATS domains by targeting a unique π-π-π stacking in the YEATS-Kcr recognition with expanded π systems to maximize the stacking interaction. The inhibitors carrying a 2-furancarbonyl (e.g., XL-07 and XL-07i) and 5-oxazolecarbonyl (e.g., XL-13 and XL-13a) at lysine side chain bound to AF9 and ENL YEATS domains with largely enhanced binding affinities (Fig. 2 and 4). Although π stacking is commonly regarded as a relatively weak non-covalent interaction, it is frequently present at ‘hot spots’ of protein interfaces37,38. Our study demonstrates that targeting π stacking is a feasible strategy for the development of inhibitors against protein-protein interactions. This strategy can be readily extended to develop inhibitors for other YEATS domains including YEATS2 and GAS41, whose biological significance as Kac/Kcr ‘readers’ remains poorly understood.
The YEATS domains of AF9 and ENL share a high structural similarity. In this study, we developed the selective inhibitors XL-13a and XL-13m for AF9 and ENL YEATS domains, respectively. While both inhibitors carry oxazolecarbonylated lysine that would occupy the proteins’ Kac/Kcr-binding pockets, there are subtle differences in the sequences and lengths of the peptide-based inhibitors’ N-terminal fragments. Since the N-terminal fragments of the inhibitors extend to the L8 loop region of the YEATS domains (Fig. 3), the differences in the protein-inhibitor interactions at this region may account for the inhibitors’ selectivity toward AF9 and ENL. Interestingly, a recent study identified the mutations in the L8 loop of ENL YEATS in Wilms tumor39, an embryonal neoplasm of the childhood kidney. The mutations were shown to alter the binding affinities of the ENL YEATS toward histone H3K9/27ac marks. Further characterizations suggested that the ENL YEATS mutations play an important role in the etiology of a subset of Wilms tumor. Based on our current study, it is highly possible that inhibitors that selectively target the ENL mutants in Wilms tumor can be developed by varying the residues in contact with the L8 loop. Such inhibitors would be useful to probe the roles of ENL during early renal development and pathogenesis of Wilms tumor.
The epigenetic landscape of a cell is commonly orchestrated by combinatory actions of multiple epigenetic proteins40,41. In MLL-translocated leukemias, the histone methyltransferase DOT1L is commonly recruited by the MLL-fusion proteins (e.g., MLL-AF4 or MLL-AF9), leading to abnormal active transcription of multiple oncogenes by installing the H3K79 methylation marks at the targeted loci42–44. These studies demonstrated the potential of DOT1L inhibition in the disruption of leukemogenic transcription programs. At the same time, histone methylation and acetylation are known to interplay in regulating malignant transcriptional programs. For example, DOT1L, via catalyzing H3K79 methylation, facilitates the recruitment of the acetyltransferase EP300, which in turn acetylates the lysine residues on the N-terminal tail of histone H4 (e.g., H4K5) and enables the binding of BRD4 to chromatin for activation of gene expression at critical MLL-fusion protein-driven oncogenes45. These studies demonstrated the therapeutic potential of combinatorial modulation of histone methylation and acetylation against MLL-translocated leukemias. In recent studies, the functional cooperation between ENL and BRD4 or DOT1L was also observed in the regulation of leukemogenic transcription programs24,25. suggesting that ENL YEATS inhibition is likely a new therapeutic strategy for the treatment of acute leukemia. In this study, we demonstrated that exposure of cells to XL-13m, an ENL YEATS-selective inhibitor, diminished the occupation of ENL on a series of genes related to leukemia initiation and suppressed their transcription. We further showed that XL-13m synergized with BET and DOT1L inhibitors, leading to the enhanced down-regulation of a set of genes that were essential for leukemogenesis and leukemia maintenance. Therefore, evaluation of the anti-leukemia activity of XL-13m and other developed ENL-inhibitors will be the important next step. While the commonly poor cell permeability of peptide-based inhibitors may hinder their application in cell-based investigations, such limitation can be overcome by a couple of strategies, such as encapsulation of inhibitors using liposomes46.
The inhibitors developed in this study also open the opportunity for the investigations of unknown roles played by ENL through a YEATS-dependent way. ENL, or its paralog AF9, is a key component of the SEC, a multiprotein complex involved in the regulation of rapid transcription induction47. A study on the SEC-dependent transcription revealed that the YEATS domain targeted the SEC to chromatin through direct binding with the RNA polymerase II (Pol II)-associated factor complex (PAFc)48. It is therefore interesting to examine whether the YEATS-PAFc interaction could be dependent on the capability of YEATS to recognize Kac/Kcr marks on the PAFc. In a recent study, histone H3K9ac was found to mediate the recruitment of SEC onto chromatin to facilitate the release of paused Pol II at selected gene promoters49. Although a solid evidence is lacking, it is likely that the YEATS domain, as a H3K9ac ‘reader’, is responsible for the SEC recruitment. We anticipate that the use of the inhibitors developed in this study would provide new evidences to dissect the mechanisms underlying the recruitment of SEC onto chromatin.
ONLINE METHODS
General.
Photo-cross-linking were performed with ENF-260C/FE hand-hang UV lamp (Spectroline). In-gel fluorescence scanning was performed using a Typhoon 9410 variable mode imager from GE Healthcare Life Sciences (excitation 532 nm, emission 580 nm). Immunoblotting membranes were visualized by MyECL Imager from Thermo Scientific. All images were processed by ImageJ software (National Institutes of Health), and contrast was adjusted appropriately. ITC experiments were performed with a MicroCal iTC200 titration calorimeter (MicroCal). IC50 and dissociation constants Kd were fit with Origin 7.0 software package (OriginLab).
Reagents and antibodies.
Unless otherwise noted, all the chemical reagents were purchased from Sigma-Aldrich and used without further purification. Antibodies used include anti-Flag (Sigma-Aldrich; F3165), anti-ENL (Bethyl Laboratories; A302–267), anti-AF9 (Bethyl Laboratories; A300–596), anti-γ-Actin (Santa Cruz; sc-65636), and goat anti-rabbit IgG-HRP (Santa Cruz; sc-2004).
Synthesis of Fmoc-protected amino acids and peptides.
Synthetic routes and spectroscopic characterizations of each compound are described in Supplementary Note.
Protein expression and purification.
Plasmids construction, protein expression and purification of AF9 (1–138), ENL (1–138), YEATS2 (201–332), GAS41 (15–159), Sirt3 (102–399), full length SPIN1, and ING2 (208–270) were performed as described previously17,50,51.
Plasmid for E. coli expression of the second BrD of BRD4 was generous gift from Dr. H. Sun (City University of Hong Kong). Complementary DNA encoding BrDs of CREBBP (1081–1197) and BAZ2B (2054–2168) were cloned into pET28a-derived vector. All BrDs were expressed in E. coli Rosetta (DE3) (Novagen) as His6-tagged fusions. Protein expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM when OD600 reached 0.6, and the cultures were further grown for 16–20 h at 18 °C. Cells were harvested and resuspended in lysis buffer (20 mM Tris–HCl, pH 8.0, 250 mM NaCl, 5% glycerol, 10 mM imidazole, 1 mM PMSF, and 1× Roche Complete EDTA-free protease inhibitors). Following sonication and centrifugation, the supernatant was loaded onto a nickel column pre-equilibrated with lysis buffer. The column was washed with 5 column volumes of wash buffer (lysis buffer with 30 mM imidazole) and then the target proteins were eluted with elution buffer (lysis buffer with 250 mM imidazole). The proteins were further purified by a Superdex75 gel filtration column. After concentration, the target proteins were frozen and stored at −80 °C.
Cell culture.
MV4;11 cell line is a gift from Prof. Anskar Yu-Hung Leung (The University of Hong Kong). HeLa S3, HEK 293T, and HEL cells were purchased from American Type Culture Collection (ATCC). MOLM-13 cells stably expressing Flag-tagged ENL was generated as previously described24. HeLa S3 and HEK 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. MOLM-13 (Flag-ENL), MV4;11 and HEL cells were cultured in RPMI 1640 medium with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Blasticidin (10 μg/mL) was further supplied into MOLM-13 (Flag-ENL) cell culture medium to maintain the expression of Flag-ENL. Cells were maintained in a humidified 37 °C incubator with 5% CO2.
Preparation of nuclear extracts.
The harvested cells were suspended with buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, and 10 mM KCl. Freshly added 0.5 mM PMSF and 1× Roche Complete EDTA-free protease inhibitors). Homogenized the suspension with Dounce homogenizer (B type pestle, 20 strokes) and incubated on ice for 10 min. Centrifuged for 15 min at 800×g at 4 °C. The supernatant was discarded. The pellet was further centrifuged for 15 min at 21,000×g at 4 °C. The supernatant was discarded. The resulting pellet was resuspended with buffer B (20 mM HEPES, pH 7.9, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, and 0.2 mM EDTA. Freshly added 0.5 mM PMSF and 1× Roche Complete EDTA-free protease inhibitors.) Homogenized the suspension with Dounce homogenizer (B type pestle, 20 strokes) and incubated on ice for 30 min on ice. After centrifugation at 21,000×g for 30 min at 4 °C, the supernatant was collected as nuclear extracts and dialyzed in binding buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.1% tween-20, 20% glycerol, and 0.5 mM PMSF) overnight at 4 °C. Protein concentration was measured by Bradford (Bio-Rad).
Photo-cross-linking.
The Probes in the absence or presence of different concentrations of competitors were incubated with recombinant proteins (5 or 20 ng/μL) or nuclear extracts (1 mg/mL) in binding buffer (50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, 0.1% Tween-20, 20% glycerol, pH 7.5, with or without 50 ng/μL BSA) for 10 min at 4 °C. Then, the samples were irradiated at 365 nm using UV lamp for 20 min in 96-well (recombinant proteins, 75 μL each well) or 12-well plate (nuclear extracts, 1 mL each well) on ice.
Cu(I)-catalyzed azide-alkyne cycloaddition/”Click” chemistry.
To the prepared photo-cross-linking samples, 100 μM rhodamine-N3 for in-gel fluorescence (10 mM stock in DMSO) or biotin-N3 for pulldown (5 mM stock in DMSO) was added, followed by 1 mM TCEP (freshly prepared 50 mM stock in H2O), 100 μM TBTA (10 mM stock in DMSO), and finally the reactions were initiated by the addition of 1 mM CuSO4 (freshly prepared 50 mM stock in H2O). The reactions were incubated for 1 h at room temperature with regular vortexing. The reactions were quenched by adding 5 volumes of ice-cold acetone and placed at −20 °C overnight to precipitate proteins.
In-gel fluorescence scanning.
Samples after protein precipitation were centrifuged at 6000×g for 5 min at 4 °C. The supernatant was discarded and the pellet was washed with ice-cooled methanol twice and air-dried for 10 min. The proteins were resuspended in 1× LDS sample loading buffer (Invitrogen) with 50 mM DTT, heated at 80 °C for 8 min. Samples were then resolved by SDS-PAGE. The labeled proteins were visualized by scanning the gel on a Typhoon 9410 variable mode imager (excitation 532 nm, emission filter 580 nm).
Isothermal titration calorimetry measurements.
Experiments were performed at 25 °C on a MicroCal iTC200 titration calorimeter (MicroCal) in buffer (150 mM NaCl, 50 mM HEPES, 1 mM TCEP, pH 7.5). The reaction cell contained 200 μL of 30−100 μM proteins, which was titrated with corresponding peptides (10-fold higher than the protein concentration used). The titration contained 17−20 injections, first 0.5 μL, and all subsequent injections of 2 μL. The binding isotherm was fit with Origin 7.0 software package (OriginLab), using a single set of independent sites to determine the thermodynamic dissociation constants and stoichiometry.
Crystallization, data collection and structure determination.
Prior to crystallization, AF9 (1–138) protein sample was mixed with XL-07i at 1:2 molar ratio for about 2 h. The complex sample was then concentrated to ~8 mg/ml ready for use. Crystallization was performed under 4 °C via vapor diffusion method under the condition: 0.2 M ammonium sulfate, 0.1 M sodium citrate tribasic dihydrate, pH 5.6 and 25 % (w/v) PEG 4,000. The crystals were briefly soaked in cryoprotectant solution containing the reservoir solution supplemented with 10% glycerol and flash-frozen in liquid nitrogen for data collection at 100 K. Diffraction data were collected at Shanghai Synchrotron Radiation Facility BL17U under cryo-conditions at 0.9791 Å. All diffraction images were indexed, integrated, and merged using the HKL2000 software package52. The structure was determined by molecular replacement using MOLREP53 with the AF9 YEATS/H3K9ac structure (PDB code: 4TMP) as the search model and refined by the program PHENIX54 with iterative model building by the program COOT55. Ramachandran Plot analysis revealed no outliers in backbone dihedral angle distribution for the refined structure; about 98.9% and 1.1% backbone dihedral angles were located to the favored and allowed regions, respectively. Detailed data collection and refinement statistics are summarized in Supplementary Table S1. Structure figures were created using the PYMOL program (http://www.pymol.org/).
In silico modeling.
The model of AF9 YEATS domain bound with XL-13a inhibitor was obtained using AF9 YEATS-XL-07i structure (PDB code: 5YYF) as the initial model and manually adjusted in COOT.
Cellular thermal shift assay (CETSA).
The CETSA was performed essentially following the published protocol31. Briefly, MOLM-13 (Flag-ENL), MV4;11, or HEL cells were incubated with 50 μM XL-13m for 12 h, and cells were washed with PBS for three times and collected. The cell pellets were resuspended in PBS with 1× Roche Complete EDTA-free protease inhibitors (approximately 3 million cells in 100 μl). The cells (100 μl per tube) were then heated at indicated temperatures for 3 min in a thermal cycler (Bio-Rad), followed by incubation at room temperature for 3 min. Samples were frozen in liquid nitrogen, and subjected to two freeze-thaw cycles to lyse the cells. The cell lysate was centrifuged at 20,000×g at 4 °C for 20 min, and the resulting supernatant were subjected to immunoblotting analysis.
Immunoblotting.
Protein samples were resolved by SDS-PAGE and transferred onto a PVDF membrane (GE Healthcare). The membrane was blocked by 5% non-fat milk in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBST) for 1 h at room temperature, and was incubated overnight with primary antibody in blocking buffer (5% bovine serum albumin in TBST) at 4 °C. The membrane was then washed by TBST for three times, and then incubated with secondary antibodies (Santa Cruz Biotechnologies) in TBST for 1 h at room temperature. After washing by TBST for three times, the proteins were visualized with SuperSignal™ west dura extended duration substrate using MyECL Imager system (Thermo Fisher Scientific).
Chromatin immunoprecipitation (ChIP).
Chromatin immunoprecipitation was performed as previously reported25 with minor modifications. Approximate 20 million MOLM-13 (Flag-ENL) cells were treated with or without 50 μM XL-13m for 24 h. Cells were then cross-linked for 10 min at room temperature, by direct addition of 1/10 volume of 10× crosslinking solution (11% formaldehyde, 50 mM HEPES pH 7.3, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) into culture medium. Cross-linking was quenched by 0.125 M glycine for 5 min at room temperature, and cells were washed three times with PBS. Cell pellets were resuspended by ice-cold ChIP lysis buffer 1 (LB1; 1 ml per 10 million cells; 50 mM HEPES pH 7.3, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Trition X-100, 1× Roche Complete EDTA-free protease inhibitors) and rotated for 20 min at 4 °. The pellet resulted from centrifugation was resuspended in ice-cold ChIP lysis buffer 2 (LB2; 1 ml per 10 million cells; 10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× Roche Complete EDTA-free protease inhibitors) and rotated for 20 min at 4 °C. After centrifugation, the pellet was suspended in ice-cold sonication buffer (0.3 ml per 10 million cells; 50 mM HEPES, pH 7.3, 140 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 1× Roche Complete EDTA-free protease inhibitors). Samples were sonicated to a chromatin ranging from 600 bp to 800 bp using a VCX750 ultrasonic processors (Sonics & Materials; 2 mm stepped microtip; 35% amplitude; 1 second on, 4 seconds off for 8 min). The chromatin was then cleared by centrifugation. For chromatin quantification, an aliquot (around 1/10 volume) of cleared chromatin was reverse cross-linked for 20 min at 95 °C and treated with 0.2 mg/mL RNase (Thermo Fisher Scientific) for 20 min 37 °C. DNA was recovered using QIAquick PCR Purification Kit (QIAGEN) and quantified by NanoDrop spectrophotometers (Thermo Fisher Scientific). The cleared chromatin was diluted by sonication buffer and 50 μg chromatin (set 5 μg chromatin as input sample) was used for one assay. Anti-FLAG® M2 magnetic beads (Sigma-Aldrich; 50 μl suspension per 50 μg chromatin) were washed three times with cold blocking buffer (5 mg/mL BSA in PBS pH 7.4) and added to chromatin samples. Immunoprecipitation was performed overnight at 4 °C. Then the beads were washed twice with ice-cold sonication buffer, once with sonication buffer supplemented with 500 mM NaCl, once with ice-cold LiCl wash buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate), and once with TE buffer supplemented with 50 mM NaCl. Beads were next resuspended in 100 μl elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) and shook for 15 min at 65 °C on a multi shaker (TOPSCIEN Instrument). The elution was then reverse cross-linked overnight at 65 °C (Input samples were treated together with elution from this step). Samples were treated with 0.2 mg/ml RNase A (Thermo Fisher Scientific) for 2 hours at 37 °C, followed with 0.2 mg/mL proteinase K (Thermo Fisher Scientific) for 30 min at 55 °C. DNA was recovered using QIAquick PCR Purification Kit (QIAGEN) for qPCR analysis.
Quantitative PCR analysis (qPCR).
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific) essentially following the user guide. RNA was quantified, and reverse transcribed into cDNA by M-MLV Reverse Transcriptase using oligo(dT)12-18 primer (Thermo Fisher Scientific). QPCR was performed using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) on an ABI StepOnePlus system. The relative mRNA level was analyzed using ΔΔCt method. The primers used are listed in the Supplementary Table 2.
Data availability and code availability.
Crystal structure data of AF9 YEATS domain bound to inhibitor XL-07i has been deposited in the Protein Data Bank (PDB) under accession code 5YYF. Other data support the findings of this study are included in the article and/or the associated supplementary files, or available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We acknowledge support from the Hong Kong Research Grants Council Collaborative Research Fund (CRF C7029-15G to X.D.L.), the Areas of Excellence Scheme (AoE/P-705/16 to X.D.L.), the General Research Fund (GRF 17126618, 17125917 and 17303114 to X.D.L.), and the Early Career Scheme (ECS) (HKU 709813P to X.D.L.). We acknowledge the National Natural Science Foundation of China (21572191 and 91753130 to X.D.L, and 31725014 to H.L.), National Key R&D Program of China (2016YFA0500700 to H.L.), National Institutes of Health (1R01CA204639-01 to C.D.A.), the Leukemia and Lymphoma Society (LLS-SCOR 7006-13 to C.D.A), and funds from The Rockefeller University (to C.D.A.). Y.L. is a Tsinghua Advanced Fellow. L.W. is a fellow of the Jane Coffin Childs Memorial Fund. We acknowledge support from Beijing Metropolis for the Beijing Novo Program (Z18111000620000 to Y.L.) and China Association for Science and Technology for the Young Elite Scientists Sponsorship Program (to T.L.). We thank the staff members at beamline BL17U1 the Shanghai Synchrotron Radiation Facility and Dr. S. Fan at Tsinghua Center for Structural Biology for their assistance in data collection and the China National Center for Protein Sciences Beijing for providing facility support. We thank H. Sun at Department of Chemistry, City University of Hong Kong for providing plasmid of the second BrD of BRD4. We thank A.Y.-H. Leung at Department of Medicine, the University of Hong Kong for providing the MV4;11 cell line.
Footnotes
Competing financial interests
X.L. and X.D.L. have filed a patent application (US Provisional Application No. 62/590,690) related to the peptide-based inhibitors reported in this manuscript.
References
- 1.Kouzarides T Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T SnapShot: Histone-modifying enzymes. Cell 131 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Patel DJ & Wang ZX Readout of Epigenetic Modifications. Annu. Rev. Biochem 82, 81–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musselman CA, Lalonde ME, Cote J & Kutateladze TG Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol 19, 1218–1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taverna SD, Li H, Ruthenburg AJ, Allis CD & Patel DJ How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol 14, 1025–1040 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenuwein T & Allis CD Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Suganuma T & Workman JL Signals and Combinatorial Functions of Histone Modifications. Annu. Rev. Biochem 80, 473–499 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Chi P, Allis CD & Wang GG Covalent histone modifications - miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhaumik SR, Smith E & Shilatifard A Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol 14, 1008–1016 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Arrowsmith CH, Bountra C, Fish PV, Lee K & Schapira M Epigenetic protein families: a new frontier for drug discovery. Nature Reviews Drug Discovery 11, 384–400 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Helin K & Dhanak D Chromatin proteins and modifications as drug targets. Nature 502, 480–488 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Cole PA Chemical probes for histone-modifying enzymes. Nature Chemical Biology 4, 590–597 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner JM, Hackanson B, Lubbert M & Jung M Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clinical Epigenetics 1(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marmorstein R & Zhou MM Writers and Readers of Histone Acetylation: Structure, Mechanism, and Inhibition. Cold Spring Harbor Perspectives in Biology 6(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippakopoulos P & Knapp S Targeting bromodomains: epigenetic readers of lysine acetylation. Nature Reviews Drug Discovery 13, 339–358 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Shortt J, Ott CJ, Johnstone RW & Bradner JE A chemical probe toolbox for dissecting the cancer epigenome. Nature Reviews Cancer 17, 160–183 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Li YY et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 159, 558–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao D, Li YY, Xiong XZ, Chen ZL & Li HT YEATS Domain-A Histone Acylation Reader in Health and Disease. Journal of Molecular Biology 429, 1994–2002 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Schulze JM, Wang AY & Kobor MS YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire 87, 65–75 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Li YY et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Molecular Cell 62, 181–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews FH et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nature Chemical Biology 12, 396–398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao D et al. YEATS2 is a selective histone crotonylation reader. Cell Research 26, 629–632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q et al. Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain. Structure 24, 1606–1612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan L et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erb MA et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi WY et al. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nature Communications 8(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YY, Zhao D, Chen ZL & Li HT YEATS domain: Linking histone crotonylation to gene regulation. Transcription-Austin 8, 9–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niphakis MJ & Cravatt BF Enzyme Inhibitor Discovery by Activity-Based Protein Profiling. Annual Review of Biochemistry, Vol 83 83, 341–377 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Li X & Kapoor TM Approach to Profile Proteins That Recognize Post-Translationally Modified Histone “Tails”. Journal of the American Chemical Society 132, 2504–2505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang TP, Liu Z & Li XD Developing diazirine-based chemical probes to identify histone modification ‘readers’ and ‘erasers’. Chemical Science 6, 1011–1017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jafari R et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nature Protocols 9, 2100–2122 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Molina DM & Nordlund P The Cellular Thermal Shift Assay: A Novel Biophysical Assay for In Situ Drug Target Engagement and Mechanistic Biomarker Studies. Annual Review of Pharmacology and Toxicology, Vol 56 56, 141–161 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Jang MK et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular Cell 19, 523–534 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Basheer F & Huntly BJP BET bromodomain inhibitors in leukemia. Experimental Hematology 43, 718–731 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Fierz B & Muir TW Chromatin as an expansive canvas for chemical biology. Nature Chemical Biology 8, 417–427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huston A, Arrowsmith CH, Knapp S & Schapira M Probing the epigenome. Nature Chemical Biology 11, 542–545 (2015). [DOI] [PubMed] [Google Scholar]
- 37.McGaughey GB, Gagne M & Rappe AK pi-stacking interactions - Alive and well in proteins. Journal of Biological Chemistry 273, 15458–15463 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Cho KI, Kim D & Lee D A feature-based approach to modeling protein-protein interaction hot spots. Nucleic Acids Res 37, 2672–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perlman EJ et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nature Communications 6(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suganuma T & Workman JL Crosstalk among Histone Modifications. Cell 135, 604–607 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Smith E & Shilatifard A The Language of Histone Crosstalk. Cell 142, 682–685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leach BI et al. Leukemia Fusion Target AF9 Is an Intrinsically Disordered Transcriptional Regulator that Recruits Multiple Partners via Coupled Folding and Binding. Structure 21, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerry J et al. MLL-AF4 Spreading Identifies Binding Sites that Are Distinct from Super-Enhancers and that Govern Sensitivity to DOT1L Inhibition in Leukemia. Cell Reports 18, 482–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuntimaddi A et al. Degree of Recruitment of DOT1L to MLL-AF9 Defines Level of H3K79 Di- and Tri-methylation on Target Genes and Transformation Potential. Cell Reports 11, 808–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilan O et al. Functional interdependence of BRD4 and DOT1L in MLL leukemia. Nature Structural & Molecular Biology 23, 673–681 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Bruce VJ & McNaughton BR Inside Job: Methods for Delivering Proteins to the Interior of Mammalian Cells. Cell Chem Biol 24, 924–934 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Luo ZJ, Lin CQ & Shilatifard A The super elongation complex (SEC) family in transcriptional control. Nature Reviews Molecular Cell Biology 13, 543–547 (2012). [DOI] [PubMed] [Google Scholar]
- 48.He NH et al. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proceedings of the National Academy of Sciences of the United States of America 108, E636–E645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gates LA et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. Journal of Biological Chemistry 292, 14456–14472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X et al. Quantitative Chemical Proteomics Approach To Identify Post-translational Modification-Mediated Protein-Protein Interactions. Journal of the American Chemical Society 134, 1982–1985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao XC et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife 3(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymology 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Vagin A & Teplyakov A Molecular replacement with MOLREP. Acta Crystallographica Section D-Structural Biology 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Structural Biology 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Structural Biology 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure data of AF9 YEATS domain bound to inhibitor XL-07i has been deposited in the Protein Data Bank (PDB) under accession code 5YYF. Other data support the findings of this study are included in the article and/or the associated supplementary files, or available from the corresponding authors upon reasonable request.