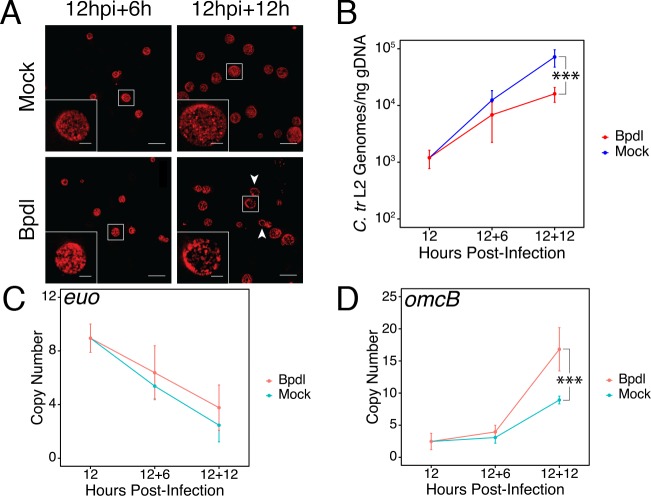
Figure 1. Brief iron limitation via 2,2-bipyridyl treatment precedes the onset of characteristic chlamydial persistence.
(A) C. trachomatis L2-infected HeLa cells were fixed and stained with convalescent human sera to image inclusion morphology by confocal microscopy following Bpdl treatment at the indicated times post-infection. Arrowheads indicate inclusions with visibly fewer organisms in the 12 hr Bpdl-treated condition. Figure shows representative experiment of three biological replicates. Scale bar = 25 μm, Inset scale bar = 5 μm. (B) Genomic DNA (gDNA) was harvested from infected HeLa cells at the indicated times post-infection under iron-replete (blue) and -depleted (red) conditions. Chlamydial genome copy number was quantified by qPCR. Chlamydial genome replication is stalled following 12 hr of Bpdl treatment, but not 6. N = 3. (C) Total RNA was harvested from infected HeLa cells at the indicated times post-infection under iron-replete (teal) and -depleted (orange) conditions. The transcript abundance of hallmark persistence genes euo and (D) omcB were quantified by RT-qPCR and normalized against genome copy number. Only at 12 hr of Bpdl treatment is omcB expression significantly affected. N = 3. Statistical significance was determined by One-Way ANOVA followed by post-hoc pairwise t-tests with Bonferroni’s correction for multiple comparisons. *=p < 0.05, **=p < 0.01, ***=p < 0.005.