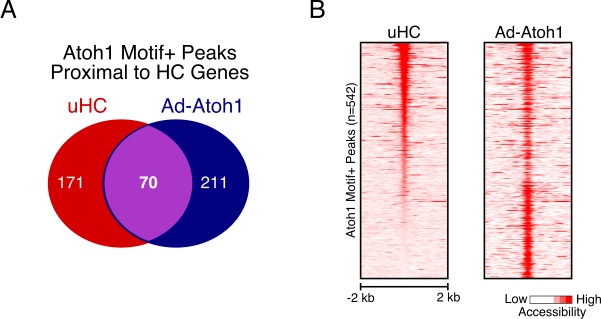
Figure 6. Atoh1 overexpression in utricle supporting cells can remodel chromatin of hair cell gene loci.
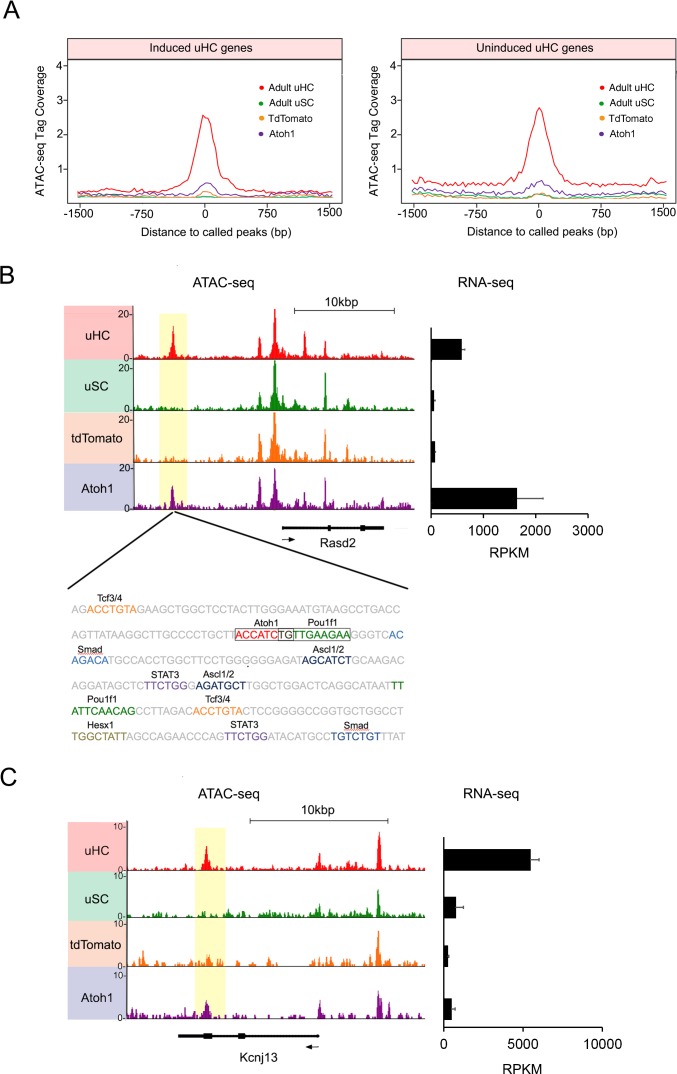
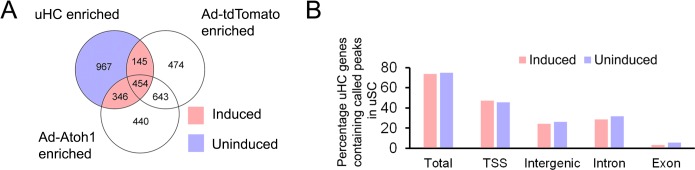
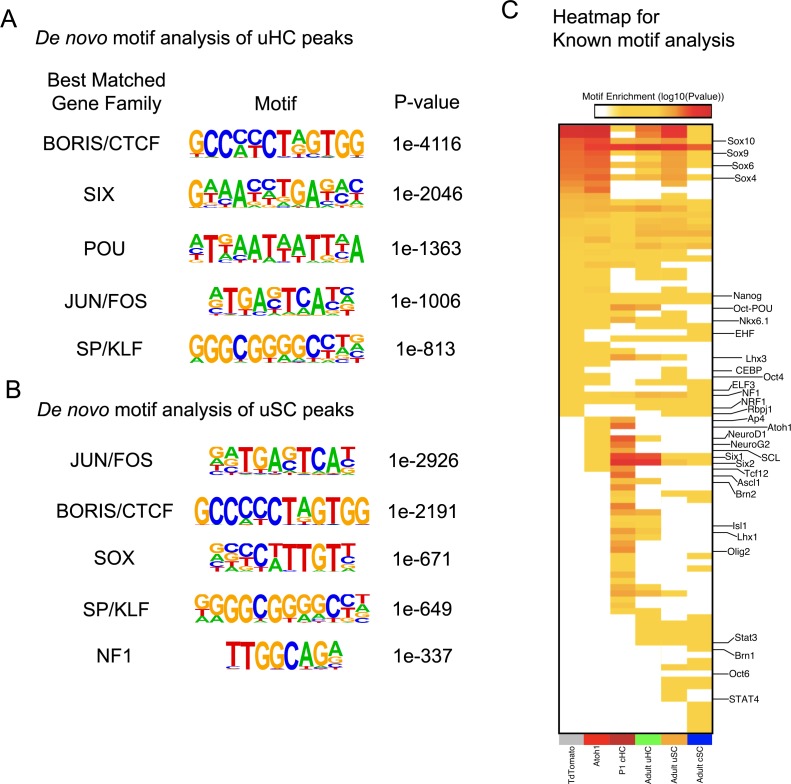
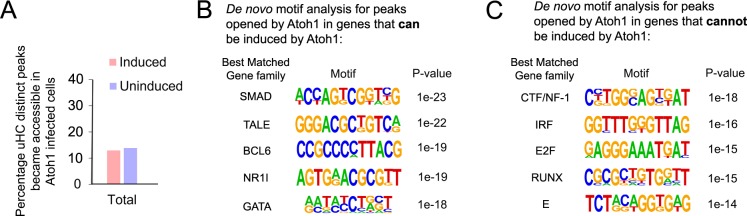
(A) Aggregation plot showing enrichment of ATAC-seq signals from each condition for unique utricle peaks that are assigned to the 945 hair cell genes that can be induced in supporting cells (left) or the 967 hair cell genes that cannot be induced in supporting cells (right). No significant differences are seen in the relative accessibility of these two groups of hair cell genes. (B) Example of an ATAC-seq profile for a hair cell-specific gene, Rasd2 that can be altered by Atoh1 transduction. RNA-seq analysis shows that Rasd2 is up-regulated in supporting cells by transduction with Atoh1, but not with the TdTomato control virus. A unique ATAC-seq peak present in hair cells but not supporting cells (yellow highlighting). However, the peak re-appears in supporting cells transduced with Atoh1, but not with the TdTomato control virus. The DNA sequence corresponding to this peak shows binding sites for Atoh1 along with other transcription factors. (C) Example of ATAC-seq profiles for a hair cell-specific gene, Kcnj13. RNA-seq analysis shows that Kcnj13is not up-regulated in supporting cells by transduction with Atoh1 or with the TdTomato control virus. Nevertheless, a unique ATAC-seq peak present in hair cells is re-opened in supporting cells transduced with Atoh1.