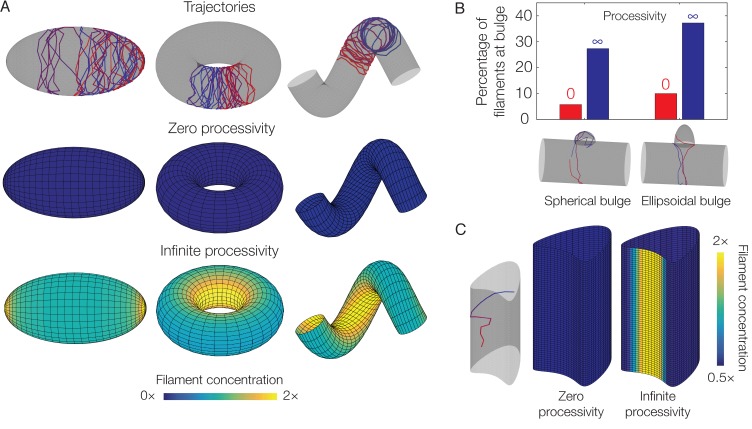
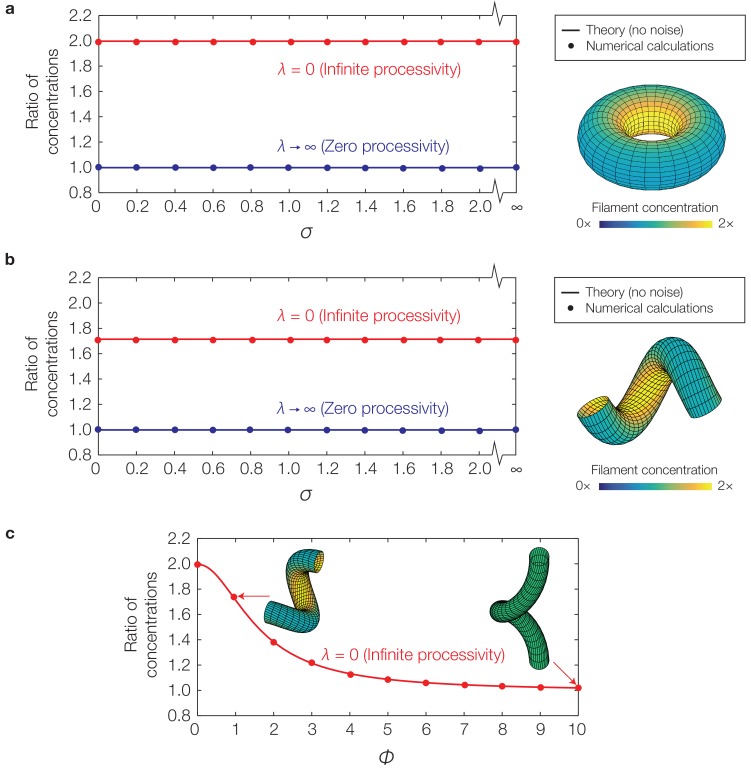
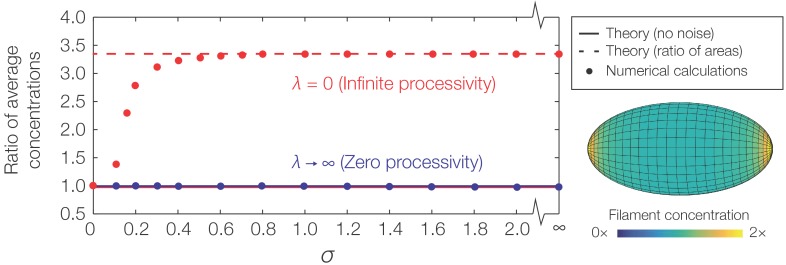
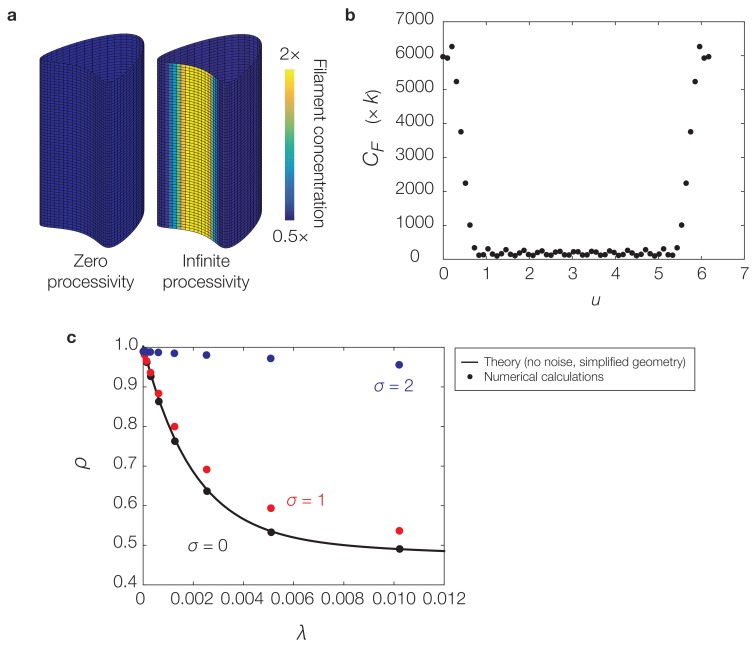
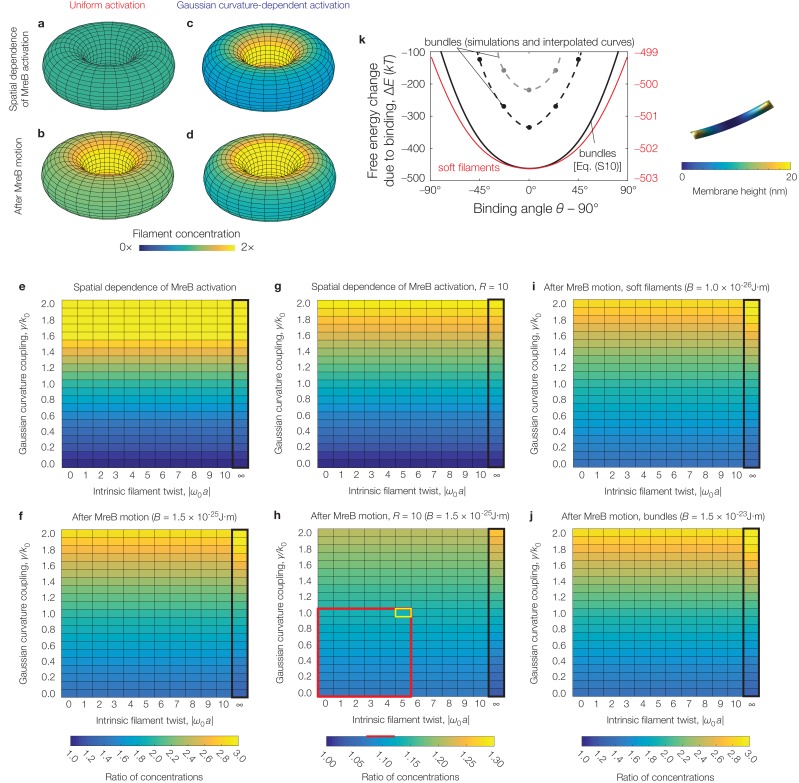
(
a-d) Numerical results for the filament concentration (
) on a torus, for the parameter values relevant to
E. coli as summarized in
Supplementary file 2 and the absence (
a, b) or presence (
c, d) of Gaussian curvature-dependent activation, with
(see also
Figure 4—figure supplement 1). (
e-j) Numerical results for the ratio of filament concentrations (see
Figure 4—figure supplement 1) for a torus across different values of intrinsic filament twist, Gaussian curvature coupling, and flexural rigidity (
). Panels (
e), (
f), (
i), and (
j), which all share the same color map, assume
while panels (
g) and (
h), which share the same color map, assume
, as to be comparable to curvatures observed in prior experiments (
Wong et al., 2017). The black box indicates parameter values where filament binding becomes energetically unfavorable. The red box highlights the characteristic range of parameter values consistent with prior experiments (
Wong et al., 2017) and estimates (
Quint et al., 2016), while the corresponding red line highlights the calculated range of concentration ratios. The cell outlined in yellow indicates values simulated in
Figure 3—figure supplement 3b. We find that the concentration ratios are largely robust to twist, but vary only for large values of Gaussian curvature coupling. (
k) Similar to
Figure 2B of the main text, but for numerical solutions of
Equation (S6) of Appendix 1 for parameters relevant to
E. coli—namely, a pressure of
—and an MreB bundle with flexural rigidity
, cross-sectional radius
, and no twist over a domain size of
(black points) and
(gray points). One such solution, corresponding to a binding angle of
for the smaller domain, is shown to the right. The numerical results are similar to the estimate of
Equation (S10) of Appendix 1 (black curve). The red curve and scale show the prediction of
Equation (S10) of Appendix 1 for a MreB filament with
and no twist.