Abstract
TP53 gene is mutated in approximately 80% of triple-negative breast cancer (TNBC). However, the prognostic significance of immunohistochemical (IHC)-detected p53 protein expression remains controversial in TNBC. In this study, we retrospectively analyzed the association between IHC-detected p53 expression and the prognosis in a cohort of 278 patients with stage I-III triple-negative breast invasive ductal carcinoma (IDC), who received surgery at the department of breast surgery in the Fourth Hospital of Hebei Medical University from 2010–01 to 2012–12. We found a positive expression ratio of IHC-detected p53 in triple-negative breast IDC of 58.6% (163/278). Furthermore, levels of expression were significantly associated with vessel tumor emboli and higher histologic grade (P = .038, P = .043, respectively), with the highest expression level observed in G3 breast cancer (64.7%). Additionally, Kaplan-Meier analysis showed that p53 expression indicated worse overall survival (OS) in the whole cohort (79.6% vs 89.6%, Log-rank test P = .025) as well as in stratified prognostic stage II patients (90.8% vs 100%, Log-rank test P = .027). The mortality risk of p53 expression patients was 2.22 times higher than that of p53 negative patients (HR: 2.222; 95%CI: 1.147–4.308). In addition, p53 expression was also associated with poor disease-free survival (DFS) (76.7% vs 86.8%, P = .020). Cox proportional hazard ratio model showed p53 expression was an independent risk factor for OS (P = .018) and DFS (P = .018) after controlling for tumor size, lymph node status, and vessel tumor emboli. Altogether, our data showed that IHC-detected p53 expression is a promising prognostic candidate for poor survival in triple-negative breast IDC patients. However, more studies are needed to determine if p53 may be applied to clinical practice as a biomarker and/or novel therapeutic target for TNBC.
Keywords: IDC, IHC, p53, progression, TNBC
1. Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-related deaths among females worldwide.[1] Triple-negative breast cancer (TNBC), accounting for approximately 15% of all breast cancers, is the most lethal subtype of breast cancer.[2] With very low or no expression of estrogen receptor (ER), progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2), TNBC is consequently unresponsive to both endocrine and anti-HER2 therapy, limiting the therapeutic option for these patients.[3] Moreover, TNBC is a heterogeneous disease in terms of clinical, histological, and molecular aspects,[4,5] Combined, these features make TNBC difficult to treat and study. To date, the molecular drivers of TNBC remain mysterious. For better treatment of TNBC, there is an urgent need to further identify and understand the signaling pathways driving TNBC tumor progression, relapse and treatment resistance.
One of the key reasons for the failure to develop a new therapy for TNBC patients is the lack of known highly prevalent, targetable molecular alteration in these tumors. Recently however, the TP53 gene was found to be mutated in approximately 80% of basal-like/TNBC,[6] raising the possibility that targeting the mutant p53 protein product might be a new approach for the treatment of TNBC. The TP53 gene, known as the “guardian of the genome”, is a tumor suppressor gene that regulates cell cycle progression, DNA repair, cellular senescence, and apoptosis.[7] Conversely, mutant p53 contributes tumor progression via a dual mechanism, the loss of tumor suppression activity and a gain of oncogenic activity.[8] As such, mutant p53 is a target of particular interest for TNBC. However, the prognostic value of p53 expression remains controversial in TNBC.[9–11] Some studies reported that breast cancers with IHC-detected p53 expression are characterized by an aggressive and metastatic phenotype with worse outcomes.[12,13] However, other studies suggested that wild-type p53 protein associated with a strong immunohistochemical signal, because the protein is commonly overexpressed as a compensatory mechanism to repair DNA damages that occur during tumorigenesis. These later studies indicated that IHC-detected p53 protein expression may serve as a favorable prognostic indicator.[14,15]
While the significance of TP53 mutation has been identified at the basic research level,[7] it remains difficult to interpret IHC results for clinical applications.[16,17] The purpose of this study is to further investigate the relationship between p53 expression in TNBC and clinical outcomes. Thus, we designed this study to evaluate IHC-detected p53 expression in a cohort of 278 patients with triple-negative breast invasive ductal carcinoma (IDC), and reveal the association between p53 expression and the prognostic impact in TNBC. Our data showed that IHC-detected p53 expression is a promising prognostic candidate for poor survival in triple-negative breast IDC patients.
2. Methods
This retrospective study comprised 278 females with stage I-III triple-negative breast IDC who underwent primary surgery at the Department of Breast Surgery in the Fourth Hospital of Hebei Medical University from 1st January 2010 to 31st December 2012. These patients underwent surgery without any anticancer therapy before the operation, including radiation therapy, systemic chemotherapy/neo-adjuvant therapy, or hormone therapy. Formalin-fixed, paraffin-embedded specimens from 278 patients were retrieved and reassessed by examining hematoxylin and eosin-stained histologic sections. The histologic type of all the specimens was reconfirmed as breast IDC, according to the World Health Organization (WHO) classification standards. Patients diagnosed with stage IV breast cancer were excluded. Additionally, patients were excluded if clinicopathological information were not available or incomplete (Fig. 1). All patients were followed up after surgery until the date of death or October 2018 (median follow-up: 79 months; range of follow-up: 2–105 months). The study was approved by the institutional ethnics committee of the Fourth Hospital of Hebei Medical University.
Figure 1.
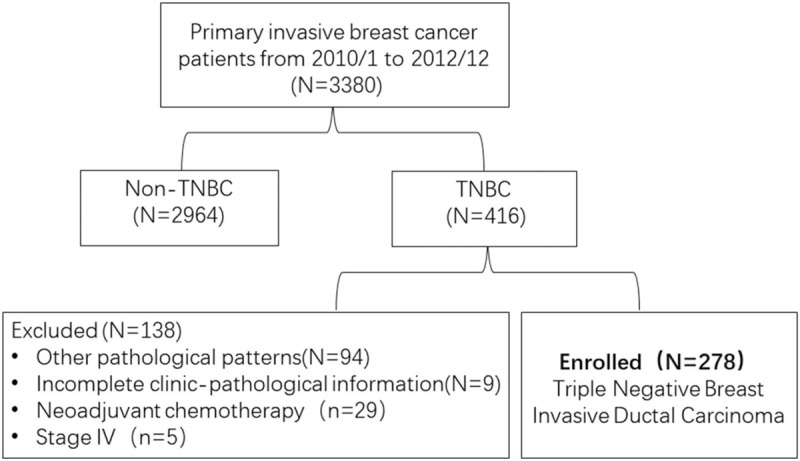
Flow diagram of the study cohort.
2.1. Histopathological determination of biomarkers
The molecular subtype of all the specimens was reconfirmed as triple-negative breast IDC by 2 experienced pathologists, according to the 14th St. Gallen International Expert Consensus. They reviewed all pathology specimens of 278 patients to determine the following tumor characteristics: histologic grade, tumor size, vessel tumor emboli and IHC staining for p53, Ki67, ER, PgR, and HER2 (all monoclonal antibodies were purchased from Abcam, Cambridge, UK). The expression of p53 is localized in the nucleus, and cells with a distribution of pale yellow or brownish yellow particles was considered p53-positive. Under the light microscope, the pathologists observed >5 high power fields with over 1000 cells and an observation of ≥10% of positive cells was considered p53 positive. Intensity of p53 was not recorded (Fig. 2).[18] Disagreements between the 2 pathologists were resolved by repeated examination of the original slide until consensus was achieved. Using a similar IHC scoring method, the Ki67 index was scored as high when 30% or more of the tumor cells were expressed.[19] Analyses for ER, PgR, and HER2 were conducted according to the recommended guidelines of the American Society of Clinical Oncology and College of American Pathologists.[20,21] Histologic grading was carried out using the Nottingham-combined histologic grade (Elston-Ellis modification of the Scarff-Bloom-Richardson grading system). Anatomic and prognostic staging was evaluated according to the 8th edition of the American Joint Committee on Cancer (AJCC) staging manual.[22] Appropriate adjuvant cytotoxic chemotherapy after the surgery were conducted according to the standard guidelines. The baseline characteristics of the 278 patients in this study are reported in Table 1.
Figure 2.
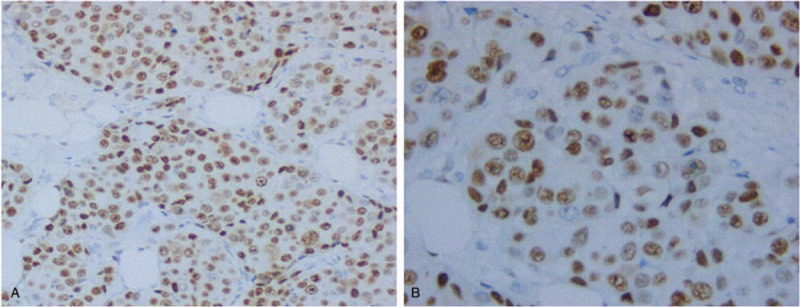
Immunohistochemical p53 staining in TNBC. Representative images of nuclear p53 expression (A, B, magnification, × 200, × 400, respectively). TNBC = triple-negative breast cancer.
Table 1.
p53 expression in TNBC according to clinicopathologic parameters.
2.2. Statistical analysis
Data were processed using SPSS for Windows, version 22.0. The association between the p53 expression and pathologic characteristics was examined using either Chi square statistical test or Fisher exact test. Overall survival (OS) and disease-free survival (DFS) were calculated from the time of diagnosis to the time of event of interest, death from any cause, recurrence, or the final follow-up date. The Kaplan-Meier method was used for calculation of survival probabilities and the unstratified log-rank test for comparison of the survival curves. Cox proportional hazards model was used to estimate the hazard ratio (HR) of each clinicopathologic variables for OS and DFS. All predictors with P value <.05 in univariate Cox analyses were used in multivariate analysis. P values were 2-tailed and considered significant when <.05.
3. Results
3.1. Patients characteristics
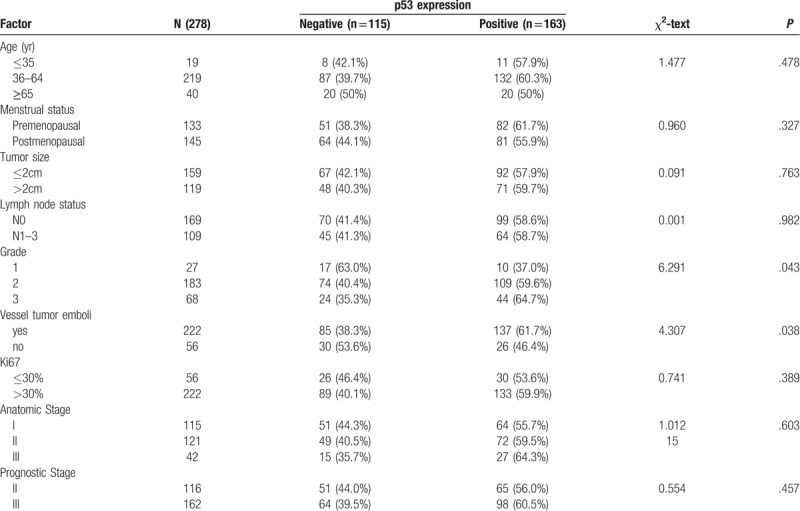
In total, 3380 patients with primary invasive breast cancer were diagnosed and treated at the Department of Breast Surgery in the Fourth Hospital of Hebei Medical University from 1st January 2010 to 31st December 2012. Among them, 416 (12.31%) patients had the TNBC subtype. After exclusion of patients with special type breast cancer (N = 94) (medullary carcinoma N = 61, metaplastic carcinoma N = 6, carcinosarcoma N = 6, secretory carcinoma N = 4, clear cell carcinoma N = 4, apocrine carcinoma N = 3, spindle cell carcinoma N = 3, papillary carcinoma N = 2, lobular breast cancer N = 1, neuroendocrine tumor N = 1, mixed carcinoma N = 3), incomplete information (N = 9) and stage IV breast cancer (N = 5), 278 patients with triple-negative breast IDC were enrolled in this study. All patients were female with a median age of 54 years (range, 23 to 80). About half (159/278) of all the patients presented with stage T1 tumor and 38.13% (109/278) had clinically detectable axillary lymph node metastasis. Around one-fifth of the patients presented with vessel tumor embolus, and 90.29% (251/278) with high histologic grade (G2 and G3). The median follow-up of the 278 patients was 79 months (range, 2 to 105), 46 patients died at data cut-off, 6 patients with local recurrence and/or distant metastasis, and the 7-year OS and DFS rates were 83.45% and 81.29%, respectively. Table 1 provides the clinicopathological characteristics of patients.
3.2. p53 expression and clinical features
The expression ratio of p53 in 278 specimens was 58.6% (163/278). There was no statistical difference between p53 (+) and p53 (−) in T stage, N stage and Ki67 expression. However, p53 expression was associated with vessel tumor embolus (61.7% vs 41.6%, P = .038) and higher histologic grade (G1 vs G2 vs G3, 37.0% vs 59.6% vs 64.7%, respectively; P = .043) Significantly, p53 expression was highest in the G3 group (64.7%).
3.3. p53 expression and survival analysis
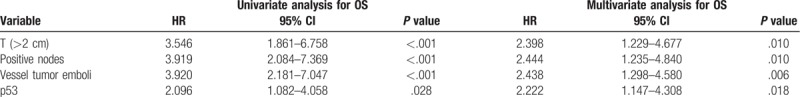
In the whole cohort of patients, Kaplan-Meier curve showed that p53 (+) TNBC conferred a significantly lower rate of DFS (76.7% vs 86.8%, log-rank P = .020, Fig. 3A) and OS (79.6% vs 89.6%, log-rank P = .025, Fig. 4A). After staging by the 8th edition of AJCC staging system, 116 had prognostic stage II cancer and 162 had prognostic stage III cancer. It showed that p53 (+) TNBC was associated with a significantly poorer OS (90.8% vs 100%, Log-rank P = .027, Fig. 4B) in the prognostic stage II group. Univariate Cox analysis of clinicopathological characteristics indicated that p53 (+) was significantly associated with worse DFS (HR: 2.036; 95%CI: 1.103–3.758; P = .023; Table 2) and OS (HR: 2.096; 95%CI: 1.082–4.058; P = .028; Table 3). Multivariate Cox analysis of all the predictors with P values < .05 in univariate Cox analyses indicated that p53 (+) was an independent prognostic factor for lower DFS (HR: 2.222; 95%CI: 1.147–4.308; P = .018, Table 2) and OS (HR: 2.097; 95%CI: 1.136–3.873; P = .018, Table 3). In addition, the multivariate analysis showed that vessel tumor emboli, T > 2 cm and positive nodes also appeared to be independent risk factors for poor prognosis (Tables 2 and 3).
Figure 3.
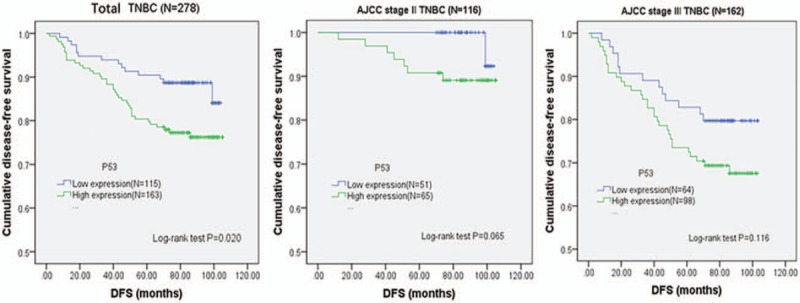
p53 prognostic significance. Kaplan-Meier curves for DFS associated with p53 expression in the whole cohort of triple-negative breast invasive ductal carcinoma patients as well as in different stratified prognostic stage patients according to the AJCC 8th edition. (A) Total TNBC, (B) AJCC prognostic stage II TNBC, (C) AJCC prognostic stage III TNBC. P values were calculated with use of the log-rank test. AJCC = American Joint Committee on Cancer, DFS = disease-free survival, TNBC = triple-negative breast cancer.
Figure 4.
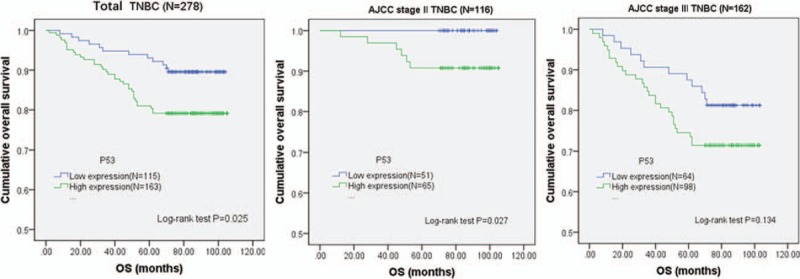
p53 prognostic significance. Kaplan-Meier curves for OS associated with p53 expression in the whole cohort of triple-negative breast invasive ductal carcinoma patients as well as in different stratified prognostic stage patients according to the AJCC 8th edition. (A) Total TNBC, (B) AJCC prognostic stage II TNBC, (C) AJCC prognostic stage III TNBC. P values were calculated with use of the log-rank test. AJCC = American Joint Committee on Cancer, TNBC = triple-negative breast cancer.
Table 2.
Univariate and multivariate analyses (Cox regression) for DFS.
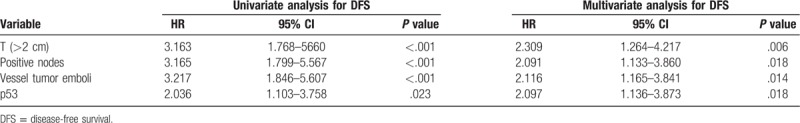
Table 3.
Univariate and multivariate analyses (Cox regression) for OS.
4. Discussion
The TP53 gene, known as the “guardian of the genome”, was found to be mutated in approximately 50% of human cancer.[23] Mutations of the tumor suppressor TP53 are associated with increased tumor cell invasion and metastasis. Several mechanisms have been identified for promoting dissemination of cancer cells with TP53 mutations.[24,25]TP53 mutation comprised 80% of TNBC, raising the possibility that targeting the mutant p53 protein product might be a new approach for the treatment of this form of breast cancer.[6] It has recently been suggested that anti-p53 drugs could reactivate mutant p53 and restore the tumor suppression activity in a preclinical model.[26–28] It is important to select appropriate patients with TP53 mutation to use these drugs and ensure successful results in clinical studies. Accurate detection of TP53 mutation would be a necessary precondition of anti-p53 therapies. Although next generation sequencing (NGS) allows whole genomic evaluation, it is still impractical in most clinical laboratories to use NGS to determine p53 mutational state due to high cost and complex interpretation. By contrast, IHC is still considered a valuable diagnostic tool with easy accessibility and well-established efficacy in investigating target agents and predicting clinical outcome in cancer. It also overcomes discrepancies between mRNA and protein expression by transcriptional and translational regulation.
IHC has provided enormous benefits in evaluating prognostic and predictive markers in cancer. Yet until now, the prognostic significance of IHC-detected p53 protein remains controversial in TNBC. According to Jin et al,[14] increased IHC expression of p53 reflected the accumulation of wild-type p53 as a compensatory mechanism of cell's DNA damage and repair system. They demonstrated that strong p53 expression served as a favorable prognostic indicator for better DFS and OS (P = .012, P = .008) in 305 TNBC cases, but not an independent prognostic factor in the multivariate analysis (P > .05). Similarly, Coates et al[16] observed that p53 expression was associated with a good prognosis in ER (−) node-negative breast cancer. However, more scholars thought that p53 protein translated from the wild-type TP53 gene was typically not observed by IHC staining with a truncated and unstable structure. Many studies showed that most IHC-detected p53 proteins are thought to be mutant p53, which are less susceptible to degradation than wild-type p53.[9–11,29] Kim et al[29] suggested that patients with TP53 missense mutations showed high protein expression in contrast to patients with wild-type TP53 (positivity of IHC: wild type vs missense, 53.6% vs 89.8%, respectively; P < .001). In fact, 3 quarters of TP53 mutations are missense substitutions rather than truncation/deletion mutations.[30] Previous studies reported that IHC-detected p53 reflected the mutant p53 and indicated poor prognosis in primary breast cancer.[12,31,32] Bae et al[12] found that the mortality risk of p53 expression TNBC was 1.84 times higher than that of p53 negative TNBC in the non-chemotherapy group (P = .027). Yang et al[33] also found that p53 predicted poor survival of breast cancer patients with visceral metastasis. However, there was no prognostic difference between p53 (+) TNBC and p53 (−) TNBC in some studies.[17,34]
In our research, we only choose patients with triple-negative subtype breast IDC for the study in order to achieve a more homogeneous study population. We found that p53 expression was associated with vessel tumor emboli (61.7% vs 41.6%, P = .038) and higher histologic grade (37.0% vs 59.6% vs 64.7%, P = .043) significantly, highest in the G3 group (64.7%). Vessel tumor emboli, an independent risk factor for prognosis, is caused by the dysfunctional cell-cell adhesive junctions and related to the complex epithelial-to-mesenchymal transition (EMT) and metastasis process.[35,36] EMT may acts as the initial event during tumor metastasis, which is the underlying cause of death in majority of breast cancer patients. In addition, TNBC is characterized by poor differentiation. The less differentiation the tumor, the more aggressive and poor the prognosis.[37] This is in line with our survival analysis that p53 expression was an independent prognostic factor for worse DFS and OS (HR: 2.222; 95%CI: 1.147–4.308; P = .01 appears to be 8) (HR:2.097; 95%CI:1.136–3.873; P = .018). This conclusion was similar to the study of Takenak et al[25] We shared the view that IHC-detected p53 reflected the mutant p53 and indicated poor prognosis. However, a limitation of our study is that we do not know the p53 mutational status of our patients, thus we cannot definitively confirm that we only detected mutant p53 in our IHC analyses.
In summary, we found that p53 appears to be an independent prognostic factor for worse prognosis. However, our finding is limited by our study's relatively small sample size and short follow-up duration. Clarifying the clinical value of p53 expression and developing a new therapy for TNBC requires more clinical study data. In addition, further research is necessary to provide insight into the biological and clinical implications of p53 expression in TNBC. The discovery of the functions regulated by p53 in tumor cells will provide not only the rational explanation of its role, but the proof of discovering a novel therapeutic target in vivo. The data identify p53, as a novel, independent prognostic factor, might aid in identifying subgroups of TNBC patients who are more likely to have a poor outcome and to whom specific therapies might be directed.
Acknowledgments
The authors would like to thank clinical pathologist Dr Yue-ping Liu, in the department of Pathology, the Fourth Hospital of Hebei Medical University, for her contributions to the study. We are also grateful to Dr. Pepper Schedin for critically reviewing our manuscript. The authors also would like to thank Mr. Weston Anderson, a professional editor in the U.S., for editorial assistance.
Author contributions
Conceptualization: Jing-ping Li, Sonali Jindal, Yun-jiang Liu.
Data curation: Jing-ping Li, Li-hua Zheng, Yun-jiang Liu.
Formal analysis: Jing-ping Li, Xiang-mei Zhang, Li-hua Zheng, Yun-jiang Liu.
Investigation: Jing-ping Li, Yun-jiang Liu.
Methodology: Jing-ping Li, Xiang-mei Zhang, Li-hua Zheng, Yun-jiang Liu.
Project administration: Jing-ping Li.
Resources: Jing-ping Li, Xiang-mei Zhang, Zhen-zhen Zhang.
Software: Xiang-mei Zhang.
Supervision: Jing-ping Li, Xiang-mei Zhang, Zhen-zhen Zhang, Sonali Jindal, Yun-jiang Liu.
Validation: Jing-ping Li, Xiang-mei Zhang, Zhen-zhen Zhang, Li-hua Zheng, Sonali Jindal, Yun-jiang Liu.
Visualization: Zhen-zhen Zhang, Li-hua Zheng, Sonali Jindal.
Writing – original draft: Jing-ping Li.
Writing – review & editing: Jing-ping Li, Zhen-zhen Zhang, Sonali Jindal, Yun-jiang Liu.
Footnotes
Abbreviations: AJCC = American Joint Committee on Cancer, DFS = disease-free survival, ER = estrogen receptor, HER-2 = human epidermal growth factor receptor 2, IDC = infiltrating ductal carcinoma, IHC = immunohistochemical, OS = overall survival, PgR = progesterone receptor, TNBC = triple-negative breast cancer.
The authors have no funding and conflicts of interest to disclose.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:69–90. [DOI] [PubMed] [Google Scholar]
- [2].Foulkes WD, Smith IE, Reisfilho JS. Triple-negative breast cancer. Wien Med Wochenschr 2010;160:174–81. [DOI] [PubMed] [Google Scholar]
- [3].Metzger-Filho O, Tutt A, De AE, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol 2012;30:1879–87. [DOI] [PubMed] [Google Scholar]
- [4].Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mayer IA, Abramson VG, Lehmann BD, et al. New strategies for triple-negative breast cancer--deciphering the heterogeneity. Clin Cancer Res 2014;20:782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Network TCGA. Comprehensive molecular portraits of human breast tumors. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet 2012;28:128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Silwal-Pandit L, Langerød A, Børresen-Dale AL, et al. TP53 mutations in breast and ovarian cancer. Cold Spring Harb Perspect Med 2017;7:a026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple negative breast cancers. Nature 2012;486:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Synnott NC, Murray A, Mcgowan PM, et al. Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer 2016;140:234–46. [DOI] [PubMed] [Google Scholar]
- [11].Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009;9:701–13. [DOI] [PubMed] [Google Scholar]
- [12].Bae SY, Nam SJ, Jung Y, et al. Differences in prognosis and efficacy of chemotherapy by p53 expression in triple-negative breast cancer. Breast Cancer Res Treat 2018;172:437–44. [DOI] [PubMed] [Google Scholar]
- [13].Jasar D, Smichkoska S, Kubelka K, et al. Expression of p53 protein product in triple negative breast cancers and relation with clinical and histopathological parameters. Pril (Makedon Akad Nauk Umet Odd Med Nauki) 2015;36:69–79. [PubMed] [Google Scholar]
- [14].Jin MS, Park IA, Kim JY, et al. New insight on the biological role of p53 protein as a tumor suppressor: re-evaluation of its clinical significance in triple-negative breast cancer. Tumor Biol 2016;37:11017–24. [DOI] [PubMed] [Google Scholar]
- [15].Lacroix M, Toillon RA. Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer 2006;13:293–325. [DOI] [PubMed] [Google Scholar]
- [16].Coates AS, Millar EK, O’Toole SA, et al. Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: results from IBCSG Trials VIII and IX. Breast Cancer Res 2012;14:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Silwal-Pandit L, Vollan HK, Chin SF, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res 2015;21:3569–80. [DOI] [PubMed] [Google Scholar]
- [18].Kurshumliu F, Gashi-Luci L, Kadare S, et al. Classification of patients with breast cancer according to Nottingham prognostic index highlights significant differences in immunohistochemical marker expression. World J Surg Oncol 2014;12:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yerushalmi R, Woods R, Ravdin PM, et al. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 2010;11:174–83. [DOI] [PubMed] [Google Scholar]
- [20].Hammond MEH, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med 2010;134:48–72. [DOI] [PubMed] [Google Scholar]
- [21].Wolff AC, Hammond MEH, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 2013;138:241–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Giuliano AE, Edge SB, Hortobagyi GN. Eighth Edition of the AJCC Cancer Staging Manual: breast cancer. Ann Surg Oncol 2018;25:1783–5. [DOI] [PubMed] [Google Scholar]
- [23].Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2–8. [DOI] [PubMed] [Google Scholar]
- [24].Antti Arjonen, Riina Kaukonen, Elina Mattila, et al. Mutant p53–associated myosin-X upregulation promotes breast cancer invasion and metastasis. J Clin Invest 2014;124:1069–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lv T, Wu X, Sun L, et al. p53-R273H upregulates neuropilin-2 to promote cell mobility and tumor metastasis. Cell Death Dis 2017;8:e2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Synnott NC, Bauer MR, Madden S, et al. Mutant p53 as a therapeutic target for the treatment of triple-negative breast cancer: preclinical investigation with the anti-p53 drug, PK11007. Cancer Lett 2018;414:99–106. [DOI] [PubMed] [Google Scholar]
- [27].Duffy MJ, Synnott NC, Crown J. Mutant p53 in breast cancer: potential as a therapeutic target and biomarker. Breast Cancer Res Treat 2018;170:213–9. [DOI] [PubMed] [Google Scholar]
- [28].Synnott NC, Murray A, Mcgowan PM, et al. Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer 2017;140:234–46. [DOI] [PubMed] [Google Scholar]
- [29].Kim JY, Park K, Jung HH, et al. Association between mutation and expression of TP53 as a potential prognostic marker of triple-negative breast cancer. Cancer Res Treat 2016;48:1338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2010;28:622–9. [DOI] [PubMed] [Google Scholar]
- [31].Eikesdal HP, Knappskog S, Aas T, et al. TP53 status predicts long-term survival in locally advanced breast cancer after primary chemotherapy. Acta Oncol 2014;53:1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Maeda T, Nakanishi Y, Hirotani Y, et al. Immunohistochemical co-expression status of cytokeratin 5/6, androgen receptor, and p53 as prognostic factors of adjuvant chemotherapy for triple negative breast cancer. Med Mol Morphol 2016;49:1–1. [DOI] [PubMed] [Google Scholar]
- [33].Yang P, Du CW, Kwan M, et al. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci Rep 2013;3:2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang J, Zhang C, Chen K, et al. ER (1 inversely correlates with PTEN/PI3K/AKT pathway and predicts a favorable prognosis in triple-negative breast cancer. Breast Cancer Res Treat 2015;152:255–69. [DOI] [PubMed] [Google Scholar]
- [35].Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer. Nat Rev Cancer 2018;18:128–34. [DOI] [PubMed] [Google Scholar]
- [36].Felipe LJ, Nofech-Mozes S, Bayani J, et al. EMT in breast carcinoma - a review. J Clin Med 2016;5:ii:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li J, Chen Z, Su K, et al. Clinicopathological classification and traditional prognostic indicators of breast cancer. Int J Clin Exp Pathol 2015;8:8500–5. [PMC free article] [PubMed] [Google Scholar]


