Abstract
The PAR-1-MARK pathway controls cell polarity through the phosphorylation of microtubule-associated proteins. The Rho-Rac guanine nucleotide exchange factor 2 (ARHGEF2), which activates the ras homolog family member A (RHOA), is anchored to the microtubule network and sequestered in an inhibited state by binding to dynein light chain Tctex-1 type 1 (DYNLT1). We showed in mammalian cells that the liver kinase B1 (LKB1) activated the microtubule affinity regulating kinase 3 (MARK3), which in turn phosphorylated ARHGEF2 at a regulatory site (Ser151). This modification disrupted the interaction between ARHGEF2 and DYNLT1 by creating a 14-3-3 binding site in ARHGEF2, thus triggering dissociation of ARHGEF2 from microtubules. Protein phosphatase 2A (PP2A) dephosphorylated ARHGEF2 Ser151 to restore the inhibited state. ARHGEF2 phosphorylation by MARK3 induced RHOA activation and stress fiber and focal adhesion formation and was required for organized cellular architecture in three-dimensional culture. We have identified a regulatory switch controlled by MARK3 that couples the microtubule and actin cytoskeletons to establish epithelial cell polarity through ARHGEF2.
Introduction
Control of cell polarity is essential for the establishment of multicellular tissues in metazoans. Genetic studies in the nematode Caenorhabditis elegans have identified a set of six partition-defective or PAR genes that participate in the polarity program during embryonic development and are conserved in mammals (1–4). PAR-1 is required for axis formation in oogenesis and establishment of oocytes in the fruit fly Drosophila melanogaster, both of which are processes associated with microtubule dynamics and stability (5). Mammals have four PAR-1 orthologs comprising the family of microtubule affinity-regulating kinases (MARKs), which are related to AMP-activated protein kinase (AMPK). The MARK family comprises four members: PAR-1a (also known as MARK3 or C-TAK), PAR-1b (also known as MARK2 or EMK), PAR-1c (also known as MARK1), and PAR-1d, (also known as MARK4 or MARKL1). MARKs are known for regulating cell polarity (3) and for triggering microtubule instability by phosphorylating microtubule-associated proteins (MAPs), causing their rapid detachment from microtubules (6, 7). The best characterized family member, MARK2, has a well-established role in cell polarity. MARK2 modulates the growth of axonal projections in hippocampal neurons (8) and contributes to the formation of neurites in neuroblastoma cells (9) through phosphorylation of the microtubule-associated protein tau (MAPT, also known as TAU). This modulates microtubule plasticity, which is required for neuronal polarity and the growth of neurites (8, 9). MARK2 also phosphorylates Rab11-Family Interacting Protein 2 (FIP2), which regulates lumen polarity (10) and the activity of Catenin delta 1 (CTNND1, also known as catenin p120) at the junctional complexes (11). Loss of function of MARK2, MARK3 or MARK4 in mice leads to metabolic defects including increased metabolic rate, decreased adiposity, defective gluconeogenesis, and insulin hypersensitivity, among others (12–14). MARK2 and MARK3 can compensate for one another during embryogenesis; however, compound homozygyous knockout of both is embryonic lethal (12,15), whereas loss of three out of four alleles causes defects in the development of the glomerular and proximal tubules of the kidneys (16). All four MARK kinases are targets of the Helicobacter pylori virulence factor CagA, which disrupts tight junctions and polarity in epithelial cell lines (17). The identification of other microtubule-associated proteins which are MARK substrates directing cell polarity has yet to be fully elucidated (18–22).
The RHOA-guanine nucleotide exchange factor ARHGEF2 has been implicated in a multiplicity of cellular processes involving the establishment of cell polarity, including epithelial tight junction formation (23) proximal tubule paracellular permeability (24), and endothelial permeability (25). We recently described a RHOA-independent requirement of ARHGEF2 in rat sarcoma (RAS)-mediated transformation (26). ARHGEF2 is sequestered in an inhibited state on the microtubule array, where it is tethered by the dynein motor light chain DYNLT1 (27, 28), and phosphorylated by p21 (RAC1) activated kinase 1 (PAK1) or protein kinase A (PKA) on the C-terminal negative regulatory site Ser886 (28, 29). Phosphorylation at Ser886 creates a binding site for 14-3-3 proteins, which hold ARHGEF2 in a catalytically inactive configuration (28). ARHGEF2 can be activated by disassembly of the microtubule array using pharmacologic agents or by the physiologic ligands lysophosphatidic acid and thrombin (30).
To elucidate the detailed mechanisms by which ARHGEF2 is positively regulated and coupled to the cell polarity program we sought to systematically determine the ARHGEF2 interaction network using a proteomic approach. We identified MARK3 as a positive regulator of ARHGEF2. MARK3 phosphorylated ARHGEF2 on Ser151, which we demonstrated using X-ray crystallographic structure determination, is contained within the DYNLT1-binding region. We showed that the sequence encompassing Ser151 is conserved across species and is a functional 14-3-3 binding site. When 14-3-3 bound to phosphoSer151, ARHGEF2 was displaced from the dynein motor complex, thereby promoting a catalytically active ARHGEF2 pool that was dissociated from microtubules. Binding of 14-3-3 to phosphoSer151-ARHGEF2 induced its relocalization to the plasma membrane and cytoplasmic compartments, where it activated RHOA to promote the induction of stress fibers and focal adhesions, whereas Ser151 phosphorylation was antagonized by the PP2A phosphatase. We identified ARHGEF2 as a MARK3 substrate activated by a phosphor-switch controlling its association with DYNLT1, and showed this is required for the formation of polarized spheroid structures.
Results
ARHGEF2 interaction network comprises proteins associated with microtubules or cell division processes
To identify new ARHGEF2 interactors we used proximity-dependent biotin identification (BiolD) (31) by stably expressing Flag-tagged bifunctional ligase-repressor (BirA*) fused to ARHGEF2 in 293 Flp-In T-REx cells, followed by affinity purification and mass spectrometry (AP-MS) (32). We confirmed six previously known ARHGEF2 interacting proteins including p21 (RAC1) activated kinase 4 (PAK4), MARK2, centrosomal protein 170 (CEP170) (33–36) and the protein phosphatase 6 (PPP6) catalytic subunit (PPP6C), regulatory subunit 1 (PPP6R1) and the ankyrin repeat domain 28 (ANKRD28) subunit (37) (Table S1, Figure 1A and S1A). Gene ontology (GO) analysis and a survey of the literature revealed that many of the ARHGEF2 interactors were associated with (i) microtubule organization and regulation, (ii) vesicle-mediated transport, (38), (iii) mitotic cell cycle processes, and (iv) cell polarity (Figure S1B, Tables S2, S3). One of the ARHGEF2 interactors of particular interest was MARK3, also known as C-TAK1. MARK3 was first cloned based on its ability to associate with and phosphorylate cell division cycle 25C (Cdc25C) (39). Subsequently, MARK3 has been shown to interact with many substrates, including protein tyrosine phosphatase H1 (PTPH1), kinase suppressor of Ras1 (KSR1) and plakophilin2 (PKP2) (40). We investigated further the function of the MARK3 :ARHGEF2 complex because these proteins have a common role in promoting microtubule dynamics.
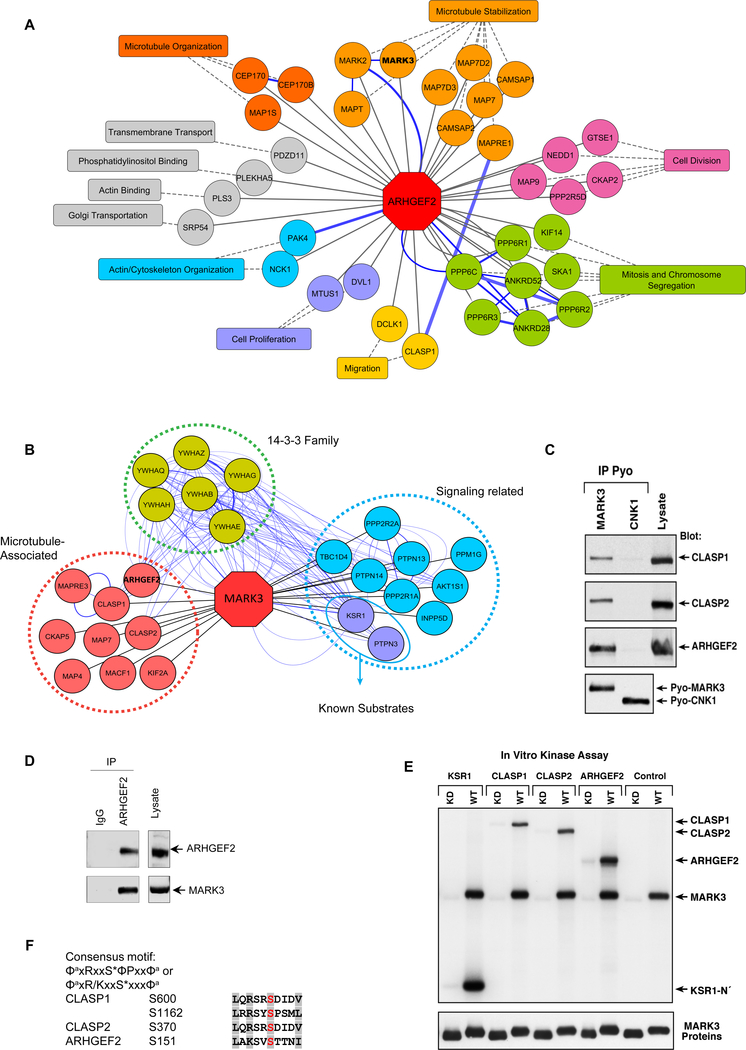
Figure 1. Interaction networks of ARHGEF2 and MARK3.
(A) ARHGEF2 high-confidence interactors detected by BioID mass spectroscopy (see Table S1). Proteins with roles in similar biological processes are grouped by the indicated functions (see Table S3), and reported protein-protein interactions (GeneMANIA) are highlighted with blue edges.
(B) MARK3 interactors detected by mass spectroscopy of immunoprecipitated Pyo-MARK3 complexes (See Table S4). Reported protein-protein interactions (GeneMANIA) are highlighted with blue edges.
(C) Pyo-tagged wild-type MARK3 and CNK1 (negative control) were co-expressed along with CLASP1, CLASP2, and ARHGEF2 and immunoprecipitated from Cos cell lysates. The protein complexes were examined by Western blot using specific antibodies for CLASP 1, CLASP2 and ARHGEF2 and Pyo for MARK3.
(D) Cell lysates from HEK293T were immunoprecipitated using IgG or an antibody recognizing ARHGEF2 combined with Sepharose beads. The protein complexes were separated by SDS page and probed with antibodies recognizing MARK3 or ARHGEF2. Whole cell lysates were analyzed by Western blots to assess MARK3 and ARHGEF2 protein abundance.
(E) KSR1 N’ (N-terminal head domain of KSR1), CLASP1, CLASP2 and ARHGEF2 proteins were immunoprecipitated from Cos cells and incubated with purified active wild type (WT) or kinase-dead (KD) MARK3 in the presence of 32P-ATP. The labeled proteins were separated by SDS-PAGE and visualized by autoradiography. The membrane was then probed for MARK3 to detect the purified MARK3 proteins.
(C-E) Representative of three independent experiments.
(F) Analysis of the protein sequences of CLASP1, CLASP2 and ARHGEF2 for the consensus MARK3 phosphorylation motifs ΦaxRxxS*ΦPxxΦa and ΦaxR/KxxS*xxxΦa using ScanProsite. (S* is the site phosphorylated, x any amino acid, Φa is a hydrophobic residue with an aliphatic side chain and Φ is any hydrophobic amino acid).
MARK 3 interacts with and phosphorylates ARHGEF2 and other microtubule-associated proteins
To understand the potential role of MARK3 and ARHGEF2 signaling networks we next sought to elucidate the protein interaction network of MARK3. Pyo-tagged MARK3 protein complexes were affinity purified from 293T cells and identified using mass spectrometry. Known MARK3 substrates including KSR1 and PTPH1 were detected in this analysis in addition to new MARK3 interacting peptides (Figure 1B and Table S4). Of these, cytoplasmic linker associated proteins 1 and 2 (CLASP1 and CLASP2), and ARHGEF2 are known microtubule-binding proteins. To confirm the interaction of MARK3 with CLASP1, CLASP2, and ARHGEF2, co-immunoprecipitation assays were performed using constructs expressing Pyo-tagged MARK3 or connector enhancer of kinase suppressor of ras 1 (Pyo-CNK1, a protein scaffold not known to bind to any of the putative MARK3 substrates (41)). CLASP1, CLASP2 and ARHGEF2 were detected in immunoprecipitates of MARK3 but not CNK1 (Figure 1C). MARK3 was identified in ARHGEF2 immunoprecipitates from 293T cells, confirming an interaction between the endogenous proteins (Figure 1D). Next, we examined whether these proteins were MARK3 substrates using an immune complex kinase assay. For in vitro kinase assays, CLASP1, CLASP2, ARHGEF2, or the N-terminal domain of KSR1 were isolated from transfected Cos-7 cell under stringent lysis conditions. The immunoprecipitated proteins were then incubated with either purified wild-type MARK3 (MARK3WT) or a kinase-deficient mutant form of MARK3 (MARK3kd) in the presence of Mg+2 and γ[32P]ATP (Figure 1E). MARK3WT exhibited autokinase activity and phosphorylated the positive control substrate, KSR1, whereas MARK3kd did not. In addition, CLASP1, CLASP2, and ARHGEF2 were strongly phosphorylated by MARK3WT but not MARK3kd (Figure 1E). These data demonstrated that CLASP1, CLASP2, and ARHGEF2 are MARK3 substrates, and all contained MARK3 consensus phosphorylation motifs ΦaxRxxS*ΦPxxΦa or ΦaxR/KxxS*xxxΦa (where S* is the site phosphorylated, x is any amino acid, Φa is a hydrophobic residue with an aliphatic side chain and Φ is any hydrophobic amino acid); (40,42, 43) at sites corresponding to CLASP1 Ser600 and Ser1162, CLASP2 Ser370 and ARHGEF2 Ser151 (Figure 1F).
MARK3 phosphorylates ARHGEF2 on Ser151 and creates a 14-3-3 binding site
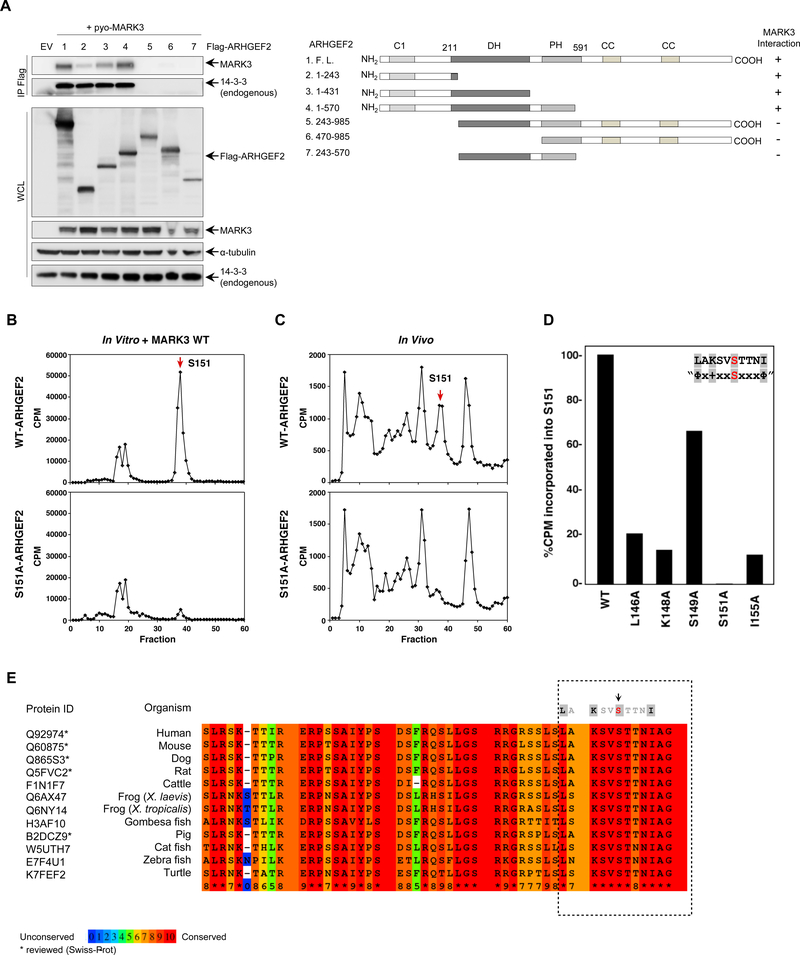
We mapped the MARK3 binding site on ARHGEF2 using a series of Flag-tagged ARHGEF2 fragments, and detected MARK3 only in immunoprecipitates containing the N-terminal portion ARHGEF2. Residues 1–243 were necessary for interaction with MARK3 and the addition of the DH and PH domains each contributed to promote maximal binding, although an isolated DH-PH domain failed to bind MARK3 (Figure 2A).
Figure 2. MARK3 binds an N-terminal region of ARHGEF2 and phosphorylates Ser151.
(A) Left: Flag-tagged ARHGEF2 fragments were co-expressed in HEK293T cells with wild-type MARK3. Protein complexes were immunoprecipitated and immunoblots were probed with antibodies specific for MARK3 and pan 14-3-3 to map the interaction. Antibodies recognizing Flag, MARK and 14-3-3 antibodies were used to detect protein abundance in cell lysates; α-tubulin was used as a loading control. Right: Schematic representation of the constructs used for mapping the interaction.
(B, C) ARHGEF2 wild-type (top panels) or mutant ARHGEF2S151A (lower panels) proteins were incubated with 32P-ATP, digested with trypsin and examined by HPLC analysis. For the in vitro analysis (B) purified MARK3 was added.
(D) Purified ARHGEF2 mutants were incubated with purified active MARK3 in the presence of 32P-ATP. The phosphorylated mutants were separated by SDS-PAGE. For each mutant the 32P-phosphate incorporated was quantitated using a phosphoimager.
(A-D) Representative of three independent experiments.
(E) Alignment of ARHGEF2 orthologs in vertebrates. The asterisks represent sequences reviewed by Swiss-Prot. The boxed area shows the consensus motif and the color code is based on the conserved residues with red being the most conserved. The full alignment is shown in Figure S2.
To determine the sites on ARHGEF2 phosphorylated by MARK3, phosphopeptide mapping analysis was performed. ARHGEF2 was phosphorylated with y[32P]ATP by MARK3 in vitro, digested with trypsin, and the phosphopeptides were analyzed by high-pressure liquid chromatography (HPLC). The major phosphorylation site was contained within a phosphopeptide that eluted in fractions 37–38. This peptide was sequenced by Edman degradation and phosphoamino acid analysis revealed that Ser151 was the residue phosphorylated in this tryptic fragment. When a similar analysis was performed using an ARHGEF2 mutant lacking Ser151 (ARHGEF2S151A), the phosphopeptide eluting in fractions 37–38 was nearly abolished (Figure 2B). Phosphopeptide analysis of in vivo 32P-labeled wildtype ARHGEF2 further confirmed that ARHGEF2 Ser151 was phosphorylated in intact cells (Figure 2C).
To validate that ARHGEF2 is an authentic substrate of MARK3, we examined whether MARK3 could phosphorylate ARHGEF2 Ser151 in vitro if key residues in the MARK3 consensus phosphorylation motif were mutated. Based on the radioactivity incorporated into Ser151, loss of the hydrophobic residues at positions −5 and +4 and loss of the lysine residue at −3 each severely reduced MARK3 phosphorylation of Ser151 (Figure 2D). These data confirmed that ARHGEF2 is a bona fide substrate of MARK3.
While ARHGEF2 orthologs exhibit 67% sequence identity overall (Figure S2), the MARK3 consensus motif residues are completely conserved among vertebrates including reptiles, amphibians, fishes and mammals suggesting an important functional role of this site (Figure 2E).
MARK3 phosphorylation of peptide sequences in other substrates frequently creates 14-3-3 binding sites. We probed the ARHGEF2 fragment comprising residues 1–243 and showed that it co-immunoprecipitated with endogenous 14-3-3 (Figure 2A). To characterize the interaction of ARHGEF2 with 14-3-3 in vitro, we analyzed the interaction between recombinant 14-3-3 protein and synthetic fluorescein isothiocyanate (FITC)-labeled peptides containing phospho-Ser151 and a previously validated 14-3-3 binding motif of ARHGEF2 at phospho-Ser885 (29). Using microscale thermophoresis (MST) we demonstrated that 14-3-3 bound the phospho-Ser151 peptide (KD of 11.6 ± 1.9 μM) and the phospho-Ser885 ARHGEF2 peptide (KD value of 2.3 ± 1.4 μM), both in a phosphorylation- dependent manner (Figure 3A).
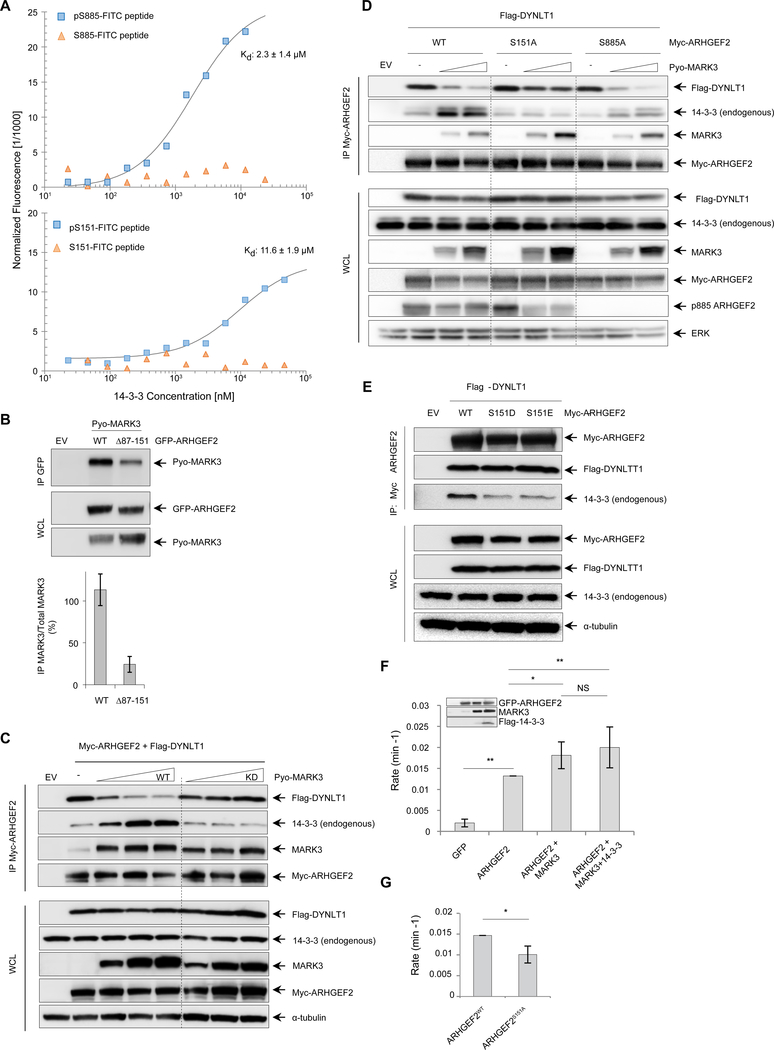
Figure 3. MARK3 perturbs the interaction between DYNLT1 and ARHGEF2 and stimulates exchange activity.
(A) Microscale thermophoresis binding assays of phosphorylated and unphosphorylated FITC-labelled ARHGEF2 peptides for Ser885 (amino acids 876–891, top panel) and Ser151 (amino acids 142–157, lower panel). The peptides were prepared at 100 nM with increasing concentrations of GST-14-3-3. Kd (dissociation constant) values were determined from the thermophoresis titration curves for the phosphorylated peptides, whereas no binding was detected for unphosphorylated peptides. Representative of three independent experiments, Kd values are average of three independent experiments ±SD.
(B) GFP-tagged wild-type ARHGEF2 and a truncated version (deletion of residues 87–151) were co-expressed with Pyo-tagged wild-type MARK3 in HEK293T cells. Protein complexes were immunoprecipitated using an antibody specific for GFP and immunoblots were probed with antibodies recognizing Pyo to detect interactions with MARK3. Antibodies specific for GFP and Pyo were used to detect protein abundance in whole cell lysates. Lower panel: quantification of the interaction normalized with total lysate. Data are means ± SD of three independent experiments.
(C) Myc-tagged ARHGEF2 was co-expressed with Flag-tagged DYNLT1 in the absence or presence of increasing amounts of wild-type (WT) or kinase-deficient (KD) MARK3. Protein complexes were immunoprecipitated using an antibody recognizing Myc antibody and analyzed by Western blot for the presence of MARK3, DYNLT1 and endogenous 14-3-3 using antibodies against MARK3, Flag and pan 14-3-3 respectively. Whole cells lysates were analyzed by Western blot and probed with the same antibodies to assess protein abundance; a-tubulin was used as a loading control. Representative of four independent experiments. See also Figure S3A, B.
(D) Myc-tagged wild-type ARHGEF2 and S151A and S885A mutants were co-expressed with Flag-tagged DYNLT1 in the absence or presence of increasing amounts of wild-type MARK3 in HEK293T cells. Protein complexes were immunoprecipitated using an antibody recognizing Myc and analyzed by Western blot for the presence of MARK3, DYNLT1 and endogenous 14-3-3 using antibodies against MARK3, Flag and 14-3-3, respectively. Whole cells lysates were analyzed with the same antibodies to assess protein abundance. Total ERK was used as a loading control. See also Figure S3C. Representative of three independent experiments.
(E) Myc-tagged wild-type and phosphomimetic mutants S151D and S151E for ARHGEF2 were co-expressed with Flag-tagged DYNLT1. Protein complexes were immunoprecipitated with an antibody recognizing Myc and analyzed by Western blot. Antibodies against Myc, Flag and 14-3-3-were used to confirm the amount of ARHGEF2 and to detect DYNLT1 and endogenous 14-3-3 in the complexes respectively. Protein abundance in whole cell lysates was analyzed using the same antibodies, and a-tubulin was used as a loading control. Representative of three independent experiments.
(F) NMR-based GEF assays were performed to measure RHOA exchange rates in the presence of cell lysates from HEK293T cells expressing GFP alone; GFP-ARHGEF2; GFP-ARHGEF2 and Pyo-MARK3; or GFP-ARHGEF2, Pyo-MARK3 and Flag-14-3-3. The amount of ARHGEF2 in exchange assays was normalized on the basis of GFP fluorescence in the lysate and protein expression concentration was detected by Western blot (inset). The rates were normalized to ARHGEF2 exchange rate. Data are means ± SD of five independent experiments. Statistical significance was determined by a Kruskal-Walis test with a Dunn’s post-test correction for multiple comparisons. *P=0.0151 (ARHGEF2 VS ARHGEF2+MARK3); **P=0.0072 (ARHGEF2 VS ARHGEF2+MARK3+14-3-3); **P=0.0079 (GFP VS ARHGEF2). NS: not significant.
(G) Nucleotide exchange rates for RHOA in the presence of cell lysates from HEK293T cells expressing GFP-ARHGEF2wt or GFP-tagged ARHGEF2S151A. The rates were normalized to ARHGEF2WT exchange rate. Data are means ± SD of four independent experiments. Statistical significance was determined by a Mann-Whitney test. *P=0.0286.
MARK3 regulates a phosphorylation-dependent switch that controls the localization and activity of ARHGEF2
We previously mapped the DYNLT1 binding site on ARHGEF2 to residues 139–161 (27). The MARK3 phosphorylation site is located within this DYNLT1 binding site, suggesting that phosphorylation, or the subsequent binding of 14-3-3 proteins, could potentially disrupt the interaction between DYNLT1 and ARHGEF2.
An ARHGEF2 mutant lacking amino acids 87 to 151 no longer binds to DYNLT1 or microtubules and is highly active (27). Deletion of these residues also diminished the interaction of MARK3 with ARHGEF2 suggesting that the MARK3 binding site overlaps with residues 87–151 (Figure 3B). Since MARK3 and DYNLT1 bind a similar region of ARHGEF2, we investigated whether MARK3 could affect the interaction between ARHGEF2 and DYNLT1. We observed that increased amounts of MARK3 potently decreased the association of ARHGEF2 with DYNLT1 (Figure 3C and Figure S3A).
We also noted that the amount of 14-3-3 in complex with ARHGEF2 tended to increase with MARK3 expression (Figure 3C and S3B). This suggested that phosphorylation of Ser151 itself, or 14-3-3 binding may be responsible for the disruption of the interface between DYNLT1 and ARHGEF2. To test if MARK3 could directly compete with DYNLT1 we expressed MARK3kd and noted no effect, demonstrating that MARK3 kinase activity is required both to increase the interaction with 14-3-3 and to disrupt the ARHGEF2:DYNLT1 interaction (Figure 3C). To probe the requirement of Ser151 to mediate the MARK3-dependent displacement of DYNLT1, we compared the interaction of DYNLT1 with ARHGEF2WT versus ARHGEF2S151A in the presence of increasing amounts of MARK3. Increasing MARK3 expression increased 14-3-3 and reduced DYNLT1 bound to wild-type ARHGEF2, but had no effect on 14-3-3 or DYNLT1 binding to the S151A mutant (Figure 3C, D and Figure S3C). By contrast, the S885A mutant exhibited less overall interaction with 14-3-3 proteins, consistent with disruption of this known binding site; however binding of 14-3-3 was increased while DYNLT1 binding was decreased by MARK3 in the same manner as wild-type (Figure 3D and Figure S3C). These data suggested that Ser151 was phosphorylated by MARK3 to create a 14-3-3 binding site that excluded binding to DYNLT1. In distinction, the C-terminal 14-3-3 binding site centered around Ser885 had no effect on DYNLT1 interaction with ARHGEF2.
To investigate whether the addition of a negative charge on Ser151 as a result of phosphorylation was sufficient to disrupt the interaction of DYNLT1 with ARHGEF2 we generated two distinct Ser151 phosphomimetic ARHGEF2 mutants by substituting the negatively charged amino acids aspartic acid (D) or glutamic acid (E) at position 151 and observed no effect on DYNLT1 binding. Both of these mutants exhibited reduced 14-3-3 binding, consistent with the recognition that 14-3-3 cannot effectively bind to phosphomimetic residues (44, 45)(Figure 3E). These results indicate that MARK3 phosphorylation promoted binding of 14-3-3 within the DYNLT1 interaction sequence of ARHGEF2 to disrupt the interaction between the dynein and the guanine exchange factor potentially affecting the microtubule-tethered inhibited state of ARHGEF2.
To determine how MARK3 might regulate the exchange factor activity of ARHGEF2 we used a real-time NMR-based assay (27,46) to measure the exchange activity in lysates of cells overexpressing MARK3. We observed that MARK3 co-expression with ARHGEF2 increased the rate of RHOA nucleotide exchange and that expression of MARK3 together with 14-3-3 further increased the exchange activity of ARHGEF2 (Figure 3F and inset). Mutation of the MARK3 Ser151 phosphorylation site on ARHGEF2 decreased its GEF activity (Figure 3G). These data support a model that MARK3 regulates a phospho-switch that leads to both the displacement of ARHGEF2 from microtubules and its concomitant activation.
ARHGEF2 Ser151 is localized in the core of the DYNLT1 binding groove
To investigate whether phosphorylation of ARHGEF2 on Ser151 directly controls the interaction between ARHGEF2 and DYNLT1 we measured the binding affinities between a non-phosphorylated FITC-labelled 16 mer-peptide derived from ARHGEF2 (residues 142–157) or a peptide phosphorylated on Ser151 to recombinant GST-tagged DYNLT1 using fluorescence polarization and determined that phosphorylation had no direct impact on binding (Figure 4A, and supported by Figure 3E). Our previous NMR characterization of the murine ARHGEF2 peptide (133–161) demonstrates that the ARHGEF2 peptide interacts with DYNLT1 with low affinity (Kd 80 μM) (27), resulting in line broadening of the resonances of both the peptide and the protein. This hindered our efforts to determine a high-resolution structure of the ARHGEF2:DYNLT1 complex by NMR, and to crystallize the complex.
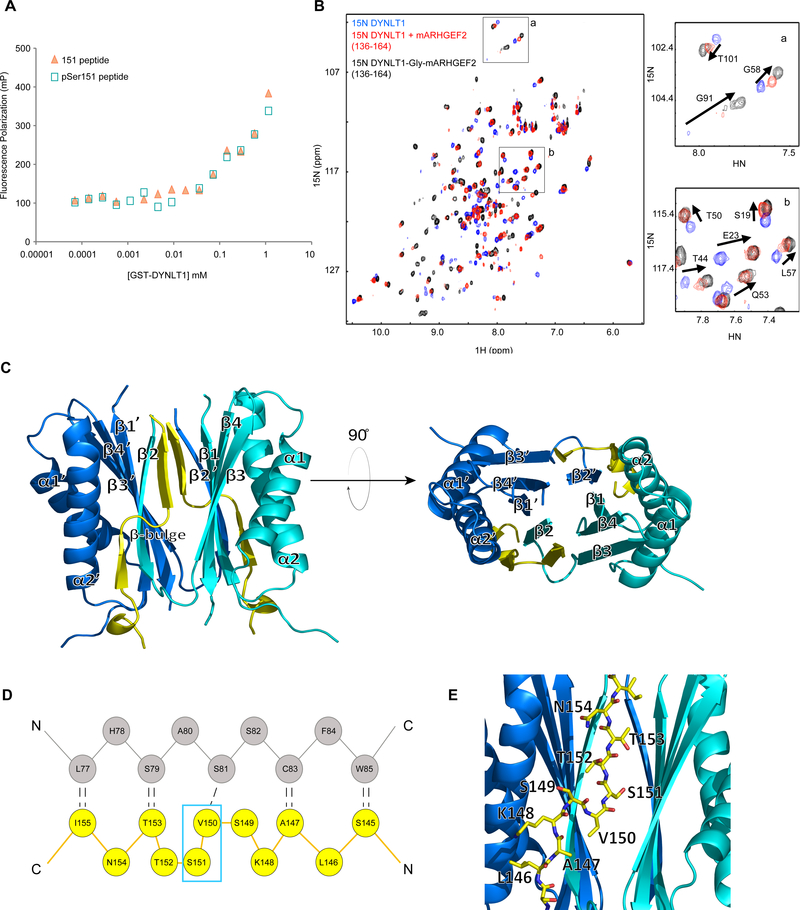
Figure 4. Structural characterization of the DYNLT1-ARHGEF2 interaction.
(A) Fluorescence polarization binding assays were performed with FITC-labelled ARHGEF2 peptides (142–157) with and without phosphorylation of Ser151. The peptides were titrated with increasing amounts of recombinant GST-DYNLT1. Representative of two independent experiments.
(B) Overlay of ‘H-15N HSQC spectra of DYNLT1 in the absence (blue) or presence (red) of mARHGEF2 peptide (136–164), or in the context of a DYNLT1-mARHGEF2 chimera with a single glycine linker (black). Inset boxes (a, b) zoom into the overlay of the spectra. Chemical shift changes for selected residues are highlighted with arrows in the spectra.
(C) Structure of DYNLT1:ARHGEF2 chimera (PDB: 5WI4). Ribbon representation, blue and cyan are used to distinguish the two subunits of DYNLT1. The ARHGEF2 portion is highlighted in yellow. See Figure S4C.
(D) Schematic representation of interactions between the domain-swapped P-strand of DYNLT1 and ARHGEF2. Residues of DYNLT1 (grey) and ARHGEF2 (yellow), with the hydrogen bond network are shown by black dotted lines. A kink in ARHGEF2 is caused by the insertion (relative to a perfect β-strand) of residues Val150 and Ser151 (cyan box), which form a β-bulge.
(E) Detail of DYNLT1 in complex with ARHGEF2. Enlargement showing the ribbon representation of DYNLT1 (blue) with a sticks model of the mARHGEF2 (yellow) component of the chimera (PDB: 5WI4). ARHGEF2 residues Leu146 to Asn154 are highlighted.
To capture the bound state of the ARHGEF2 peptide to DYNLT1 for crystallographic and NMR structural studies we coupled the ARHGEF2 (136–164) peptide to DYNLT1 using a flexible linker to enhance the apparent affinity of the interaction (47). The binding orientation of the ARHGEF2 peptide was determined using paramagnetic relaxation enhancement (PRE) experiments using a synthetic ARHGEF2 peptide (ARHGEF2-Cys137-Trp158) with an N-terminal cysteine conjugated to a maleimide-linked EDTA tag to chelate a paramagnetic ion (Figure S4A). DYNLT1 residues proximal to the N-terminus of the peptide were identified by broadening of their 15N-1H heteronuclear single quantum correlation (HSQC) cross peaks in the presence of Mn+2- versus Ca+2-bound peptide. Mapping the perturbed peaks onto the DYNLT1 structure revealed a patch near the C-terminus of DYNLT1 comprising Ser88 and Ile113 (Figure S4B), thus the ARHGEF2 peptide was fused to the C-terminus of DYNLT1, initially connected by a long flexible linker (GLEGGSGGSG) to facilitate the formation of a native interaction.
A comparison of the 15N-1H HSQC spectra of the 15N labeled chimera with that of 15N DYNLT1 in the presence of excess unlabeled ARHGEF2 (136–161) peptide validated that within the chimera ARHGEF2 bound DYNLT1 in the native manner, and saturated the site more fully (Figure 4B). Using these spectra to assess native interaction, the linker was progressively shortened (from GLEGGSGGSG, to GGSGGSG, to G) to remove unnecessary flexible regions. A single glycine linker was sufficient to maintain the native “fingerprint” spectrum, reduce peak broadening, increase saturation of the binding site, and 52% of the backbone resonances of this chimera were assigned. Whereas the apo form of murine DYNLT1 did not produce crystals despite extensive screening (~ 600 unique crystallization conditions), the chimera was readily crystallized in numerous conditions that were further optimized to generate a crystal that diffracted to high resolution.
The crystal structure of the chimera had three molecules per asymmetric unit, two of which formed a homodimer, while the third formed a dimer with a symmetrically related molecule (Figure S4C and Table S5). Similar to previously determined DYNLT1 homologue structures, each subunit had two α-helices that flanked a central β-sheet consisting of four strands (β2’,1,4,3), one of which (β2) was domain swapped from the other subunit (Figure 4C). Due to the absence of electron density, residues 1–4 and 73–76 of DYNLT1 and residues 136–138 and 156–164 of ARHGEF2 were not built into the model. ARHGEF2 residues 140–155 lay in the grooves formed at the interface of the two homodimeric DYNLT1 subunits. Residues 140–144 formed one turn of 310 helix, while residues 145–147 and 153–155 had β-strand characteristics with nine backbone hydrogen bonds formed between this stretch of the ARHGEF2 peptide (145–155) and DYNLT1 (Figure 4D). An H-bond between ARHGEF2 Ala147 and DYNLT1 Cys83 was consistent with Nuclear Overhauser Effects (NOEs) observed between these residues in solution (Figure S4D). ARHGEF2 bound to the edge of the β-sandwich, extending the domain swapped antiparallel β-sheet of DYNLT1 (Figure 4C). Relative to a perfect β-strand, ARHGEF2 contained two extra amino acids, which were accommodated in the binding site through a distortion of the β-strand (an antiparallel β-bulge comprising two type IV β-turns) (Figure 4D). The β-bulge disrupted the classic antiparallel β-sheet hydrogen bonding network (48,49): Ser151 did not participate in backbone hydrogen bonding with DYNLT1 in two crystallographically independent subunits and residues Ser149, Val150, and Ser151 exhibited large deviations from the ideal β-sheet psi/phi dihedral angles. The structure revealed that ARHGEF2 Ser151 was in the core of the DYNLT1 binding site (Figure 4E), providing a clear structural basis for the observed competition between DYNLT1 and 14-3-3 for interaction with ARHGEF2. The side chain hydroxyl of Ser151 was closely packed and formed hydrogen bonds with the hydroxyls of DYNLT1 Thr94 and Ser107 (Figure S4E), thus Ser151 phosphorylation would introduce a steric clash in this conformation. However, since our in vitro and in vivo observations indicated that phosphorylation alone or phosphomimetic mutations do not disrupt this complex, we propose that flexibility of the β-bulge may allow these residues to adopt an alternate conformation in which a phosphorylated Ser151 side chain can be accommodated in the binding groove. Several structural observations suggested that the β-bulge exhibits some conformational dynamics. Besides the hydrogen bonds mediated by the Ser151 hydroxyl, which would be spoiled upon phosphorylation, and a few bound water molecules, there were few interactions stabilizing the β-bulge, thus its conformation diverged slightly among each of the three molecules in the asymmetric unit. In NMR experiments, a stretch of four ARHGEF2 amino acids (Li44-AM7) in the chimera were assigned, however broadening of resonances from the β-bulge residues was consistent with the presence of multiple interconverting conformations. Further, phosphorylation of DYNLT1 Thr94 or phosphomimetic mutation (T94E) disrupts its interaction with dynein intermediate chain (DIC) (50), which lacks the β-bulge at this site (51), but DYNLT1T94E retains binding to ARHGEF2 (27). The DYNLT1-binding region of ARHGEF2 is remarkably rich in phosphorylation sites (Ser143, Ser149, Ser151, Thr152, Thr153, and Ser163) that have been reported in previous proteomics studies (33, 35, 52–56) and could potentially modulate this interaction. The modest affinity of DYNLT1 for monomeric ARHGEF2 peptide would allow kinases to gain access to their phosphorylation motifs, while the tethering of ARHGEF2 to microtubules is likely enhanced by avidity in multivalent protein complexes (57). Recently another mammalian DYNLT1 structure was determined in complex with the dynein intermediate chain polypeptide (58) employing this chimeric approach (59).
PP2A and MARK3 regulate the phosphorylation of ARHGEF2 in Ser151
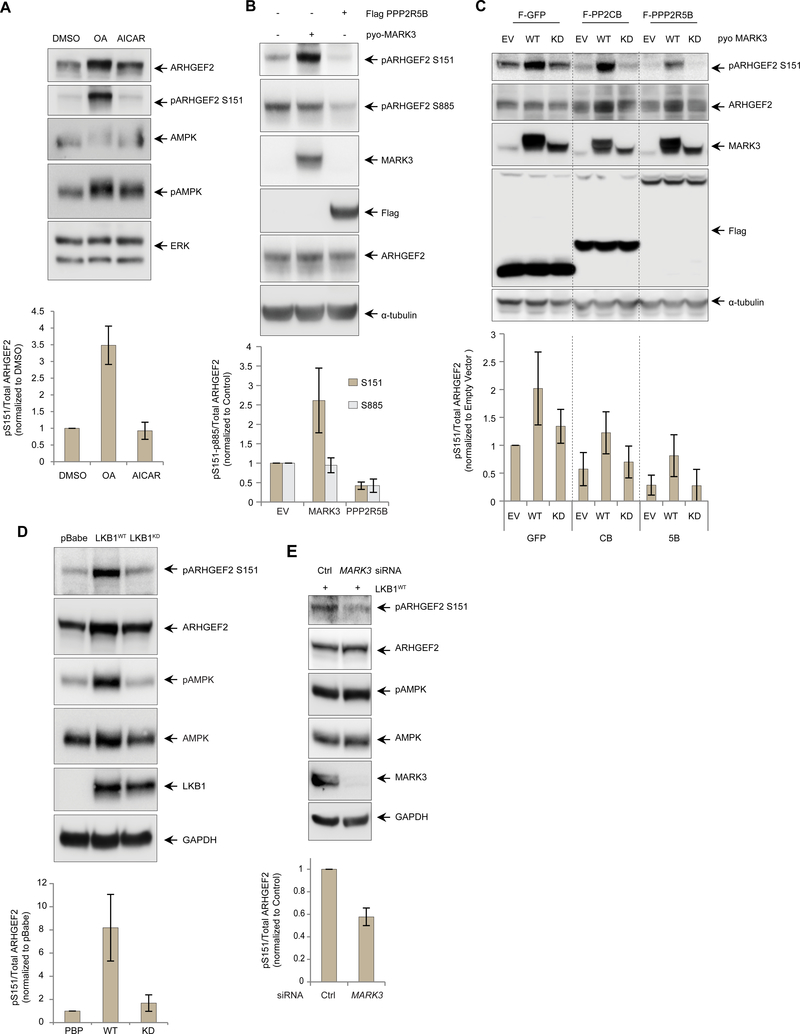
The PP2A phosphatase is a positive regulator of MAPK signaling by mediating the dephosphorylation of critical 14-3-3 binding sites in KSR1 that are phosphorylated by MARK3 (60) and it has also been described as the major phosphatase for different MAPs (61, 62). To assess the potential effect of PP2A on regulating the interaction between ARHGEF2 and DYNLT1 we treated cells with a low dose of okadaic acid (OA), an inhibitor of PP2A-family phosphatases, and evaluated Ser151 phosphorylation using phospho-Ser151-specific antibodies (Figure S5A). We observed an increase in ARHGEF2 Ser151 phosphorylation suggesting that PP2A, or a related phosphatase, dephosphorylated this residue (Figure 5A). ARHGEF2 Ser151 was recently identified in a high-throughput screen as a substrate of AMPK (56), thus we also treated the cells with the AMPK activator 5-Aminoimidazole-4-carboxamide ribonucleotide (AICAR), but no apparent change in phosphorylation of this residue was observed (Figure 5A).
Figure 5. LKB1-MARK3 axis and PP2A regulate the phosphorylation of ARHGEF2 Ser151.
(A) Western blot of HEK293T cells treated with DMSO, PP2A inhibitor okadaic acid (OA, 50 nM for 4 hours) and AMPK activator AICAR (1mM for 6 hours). Phosphorylated ARHGEF2 Ser151 was detected using a site-specific antibody and total ERK was used as a loading control (See Figure S5 for details). Lower panel: quantification of the phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independedent experiments.
(B) Western blot of HEK293T cells overexpressing pyo-tagged MARK3WT or Flag-tagged PPP2R5B. Phosphorylated ARHGEF2 Ser151 and Ser885 were detected using site-specific antibodies and a-tubulin was used as a loading control. Lower panel: quantification of Ser151 and Ser885 phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independent experiments.
(C) Western blot of 293 T-Rex Flp-In cell lines carrying inducible expression of Flag-tagged GFP, PP2A catalytic subunit PPP2CB or regulatory B’subunit PPP2R5B. The cells were induced overnight with tetracycline 500ng/ml and also transfected with empty vector, pyo-MARK3WT or pyo-MARK3KD. Phosphorylated ARHGEF2 Ser151 was detected using a site-specific antibody and α-tubulin was used as a loading control. Lower panel: quantification of the phosphorylation normalized with total ARHGEF2, Data are means ± SD of four independent experiments.
(D) Western blot of A549 LKB1-deficient cells stably expressing empty vector (pBabe), wild-type LKB1 (LKB1WT) or kinase deficient LKB1 (LKB1KD). Phosphorylation of AMPK was used as a control substrate for LKB1 phosphorylation. Phosphorylated ARHGEF2 Ser151 and AMPK Thr172 were detected using site-specific antibodies and GAPDH was used as a loading control. Lower panel: quantification of ARHGEF2 Ser151 phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independent experiments.
(E) Western blot of A549 cells expressing LBK1WT and treated with an siRNA pool specific for MARK3 versus control for 72 hours. Phosphorylation AMPK was used as a control substrate for LKB 1 phosphorylation. GAPDH was used as a loading control. Lower panel: quantification of ARHGEF2 Ser151 phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independent experiments.
To verify whether PP2A was responsible for modulating regulatory phosphosites on ARHGEF2, we overexpressed the serine-threonine-protein phosphatase 2A 56 kDa regulatory subunit beta isoform (PPP2R5B), a member of the B’ regulatory subunit of PP2A shown to specifically interact with and modulate ARHGEF2 (26, 30), in HEK293T cells and observed that both Ser151 and Ser885 were efficiently dephosphorylated (Figure 5B). We also used tetracycline-inducible cell lines expressing Flag-tagged catalytic PPP2CB and regulatory PPP2R5B subunits of PP2A, in the presence of MARK3WT or MARK3kd and observed that these PP2A subunits each tended to decrease the phosphorylation of Ser151, while co-expression of MARK3WT but not MARK3kd increased Ser151 phosphorylation (Figure 5C). No evident changes in Ser151 phosphorylation were observed when another ARHGEF2-associated phosphatase PPP6C (see Figure 1A) was overexpressed, suggesting the specificity of PP2A as a phosphatase for ARHGEF2 Ser151 (Figure S5B).
LKB1 promotes ARHGEF2 Ser151 phosphorylation through MARK3
Members of the family of MARKs have been characterized as downstream targets of the serine-threonine kinase Liver Kinase B1 (LKB1, also known as STK11 and Par-4) (63–65). LKB1 is a tumour suppressor linked to Peutz-Jeghers syndrome (66, 67), and plays a key role establishing cell polarity in Drosophila, C. elegans, and mammals (68–70). To explore a potential role of LKB 1 in regulating ARHGEF2, we probed Ser151 phosphorylation in the LKB-deficient non-small cell lung cancer cell line A549 stably expressing wild-type (LKB1WT), kinase-dead LKB1 (LKB1kd) or empty vector (pBabe), as previously described (71). ARHGEF2 phospho-Ser151 was detectable at low abundance in the control cells, but was strongly enhanced in LKB 1WT- cells. Expression of LKB 1KD had little impact on the amount of phospho-Ser151 (Figure 5D). To interrogate the role of MARK3 as the potential downstream kinase regulated by LKB 1 responsible for phosphorylation of ARHGEF2 on Ser151, we knocked down MARK3 in LKB1wt cells and observed decreased of Ser151 phosphorylation but not AMPK phosphorylation. These data provide genetic evidence that LKB 1 enhanced Ser151 phosphorylation through activation of MARK3 (Figure 5E).
MARK3 phosphorylation of ARHGEF2 triggers its redistribution from microtubules to the cytoplasm
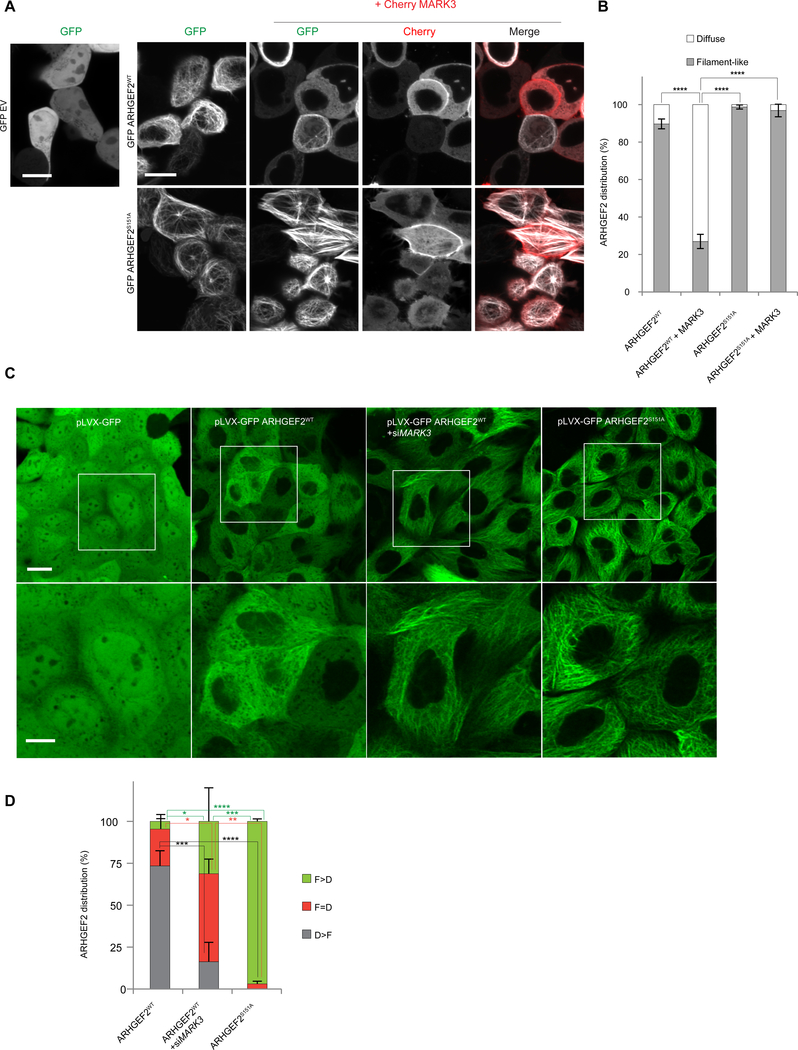
Binding of 14-3-3 proteins to the MARK3-phosphorylated substrates KSR-1, CDC25C and PKP2 is associated with alteration in their subcellular localization (40, 72, 73). We tested whether phosphorylation of Ser151 could alter the localization of ARHGEF2. GFP alone, GFP-tagged wild-type ARHGEF2 (GFP-ARHGEF2WT) or mutant S151A (GFP-ARHGEF2S151A) were over-expressed in HEK293T cells in combination with cherry-tagged MARK3 and imaged by confocal microscopy (Figure 6A). GFP-ARHGEF2WT was localized to microtubule-like filamentous structures as previously described (Figure 6A, B) (27, 28). MARK3 co-expression disrupted the filament-like localization of GFP-ARHGEF2WT, inducing a diffuse and uniform cytoplasmic distribution. In contrast, GFP-ARHGEF2S151A was strongly associated with microtubule-like filamentous structures with some bundling, which was resistant to co-expression of MARK3 (Figure 6A, B).
Figure 6. MARK3 affects the localization of ARHGEF2 dependent on Ser151 and 14-3-3.
(A, B) Live imaging pictures of HEK293T cells. In A, top panel, cells transiently overexpressing GFP alone, GFP-ARHGEF2wt alone or in combination with Cherry-MARK3; bottom panel cells expressing GFP- ARHGEF2S151A alone or co-expressed with Cherry-MARK3. ARHGEF2 distribution in A is quantified in B (as percentage of cells). A total of n=100–200 cells per condition were counted. Data are means ± SD of three independent experiments (n=3). Scale bar, 10 μm. Statistical significance was determined by a two way ANOVA test with a Bonferroni posttest correction for multiple comparisons. ****P≤0.0001.
(C, D) Live imaging of MDCKII cells stably expressing inducible pLVX-GFP, pLVX-GFP ARHGEF2WT, pLVX-GFP ARHGEF2S151A and pLVX-GFP ARHGEF2WT in combination with siMARK3; zoomed regions shown in bottom panels. In (D) quantification, as percentage of cells, of images shown in (C): cells having a higher tendency of showing a filament-like distribution (F>D); a higher tendency of having a diffuse distribution (D>F) or similar distribution of filament-like and diffuse appearing structures (D=F). F (Filament-Like distribution); D (Diffuse distribution). A total of n=250–300 cells per condition were counted. Data are means ± SD of three independent experiments. Scale bar 20 μm (upper images), 10 μm (lower images). Statistical significance was determined by a two way ANOVA test with a Bonferroni post-test correction for multiple comparisons. *P=0.0470 (in F>D ARHGEF2wt VS ARHGEF2WT+siMARK3); *P=0.0235 (in F=D ARHGEF2WT VS ARHGEF2WT+siMARK3); **P=0.0014; ***P=0.0002 (in F>D ARHGEF2WT + siMARK3 VS ARHGEF2S151A); ***P=0.0005 (in D>F ARHGEF2wt VS ARHGEF2WT+siMARK3); ****P≤0.0001.
To determine the requirement of MARK3 in determining the subcellular localization of ARHGEF2 we over-expressed ARHGEF2wt or ARHGEF2S151A at low concentration under control of a tetracycline-inducible (TETi) promoter in MDCKII cells (Figure S6A, B) (pLVX-GFP ARHGEF2WT and pLVX-GFP ARHGEF2S151A respectively). We observed a diffused distribution of pLVX-GFP ARHGEF2WT (Figure 6C). Followed by knockdown of MARK3 using siRNA (Figure S6C) we observed that a larger fraction of pLVX-GFP ARHGEF2WT was associated with microtubule-like filamentous structures in the absence of MARK3 and tended to phenocopy the subcellular filament-like distribution of pLVX-GFP ARHGEF2S151A (Figure 6C–D). These data demonstrate that MARK3-dependent phosphorylation of ARHGEF2 promoted ARHGEF2 dissociation from the filamentous structures in the cell.
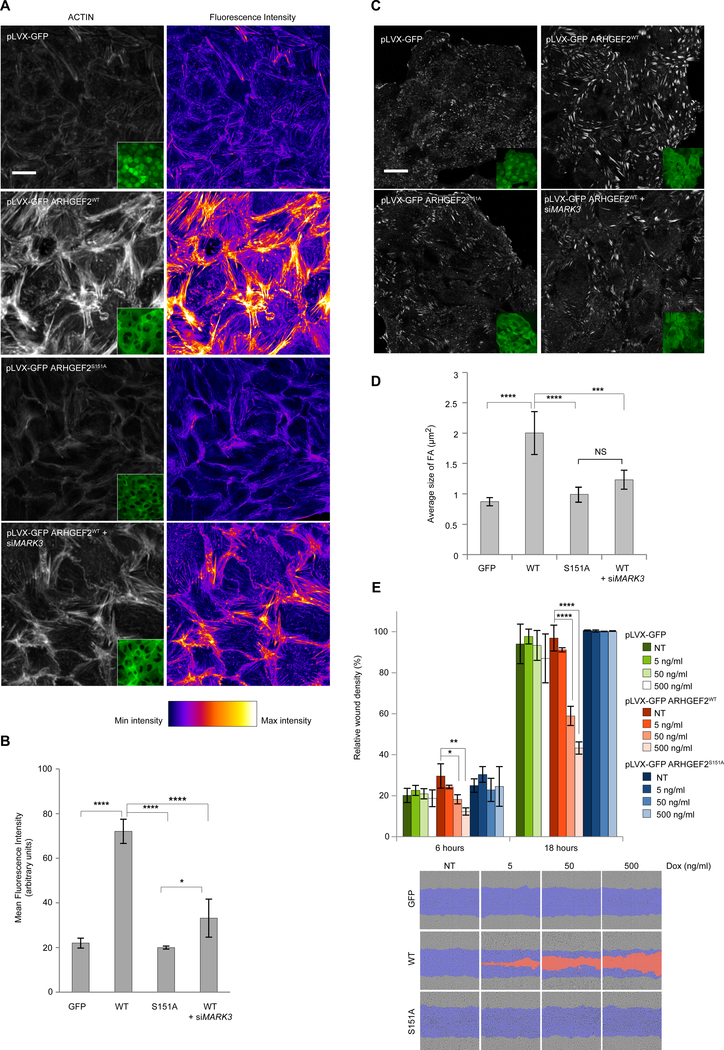
Phosphorylation of ARHGEF2 on Ser151 by MARK3 promotes stress fibers and focal adhesion formation
ARHGEF2 overexpression promotes RHOA activation and induces formation of stress fibers and focal adhesions (27). We examined the cellular effect of ARHGEF2 using the TETi system to titrate the concentrations of MARK3, ARHGEF2wt and ARHGEF2S151A (Figure S6A, S6D, E). After induction with doxycycline, the cells were fixed and stained to assess the formation of focal adhesions (VINCULIN) and stress fibers (ACTIN) (Figure 7A–D and Figure S7). Induction of pLVX-GFP ARHGEF2WT strongly enhanced the formation of stress fibers and focal adhesions and led to a decreased rate in wound closure, likely due to an enhanced adherence of the cells (Figure 7A–E and Figure S7A), whereas the induction of pLVX-GFP ARHGEF2S151A produced significantly less stress fibers and focal adhesions, and had little impact on wound healing rates compared to pLVX-GFP ARHGEF2WT (Figure 7A–E). The ability of ARHGEF2 to induce actin reorganization and focal adhesion formation was attenuated by silencing of MARK3 with specific siRNA (Figure 7A–D). Consistent with these observations, induced expression of MARK3 promoted the formation of focal adhesions and stress fibers in a manner similar to ARHGEF2 (Figure S7B–F). These data demonstrated a functional link between ARHGEF2 and MARK3, and showed that the ability of ARHGEF2 to induce polymerized actin structures was contingent on MARK3.
Figure 7. MARK3 phosphorylation of ARHGEF2 Ser151 regulates several biological functions.
(A, B) Immunofluorescence of MDCKII cells stably expressing inducible pLVX-GFP, pLVX-GFP ARHGEF2WT, pLVX-GFP ARHGEF2S151A and pLVX-GFP ARHGEF2WT in combination with siMARK3. The cells were fixed and stained for actin (left panel), GFP signal is shown (inset); right panel, LUTs showing the fluorescence intensity, white being the more intense. In B, quantification of the Mean Fluorescence intensity of the images shown in A. Five high magnification fields per condition and per experiment were quantified using ImageJ. Data are means ± SD of three independent experiments. Scale bar 20 μm. Statistical significance was determined by a one way ANOVA test with a Bonferroni post-test correction for multiple comparisons. *P=0.0453; ****P<0.0001.
(C, D) Immunofluorescence of MDCKII cells stably expressing inducible pLVX-GFP, pLVX-GFP ARHGEF2WT, pLVX-GFP ARHGEF2S151A and pLVX-GFP ARHGEF2WT in combination with siMARK3. The cells were fixed and stained for vinculin, GFP signal is shown (inset). In D, quantification of the average size of the focal adhesion (FA) of the images shown in C. Four high magnification fields per condition and per experiment were quantified using ImageJ. Data are means ± SD of three independent experiments. Scale bar, 20 μm. Statistical significance was determined by a one way ANOVA test with a Bonferroni post-test correction for multiple comparisons. ***P=0.0010; ****P≤0.0001. NS: Not significant.
(E) Relative wound density (cell density in the wound area expressed relative to the cell density outside of the wound area over time), in percentage, of MDCKII cells stably expressing inducible pLVX-GFP, pLVX-GFP ARHGEF2WTand pLVX-GFP ARHGEF2S151A exposed to increasing amounts of doxycycline at 6 and 18 hours. Bottom panel: comparison of the wounds at 18 hours. In blue, mask of the original wound, in orange, wound not closed. Data are means ± SD of three independent experiments done in triplicates. Statistical significance was determined by a two way ANOVA test with a Bonferroni post-test correction for multiple comparisons. *P=0.0445; **P=0.0029; ****P≤0.0001.
Phosphorylation of ARHGEF2 on Ser151 controls cell polarity
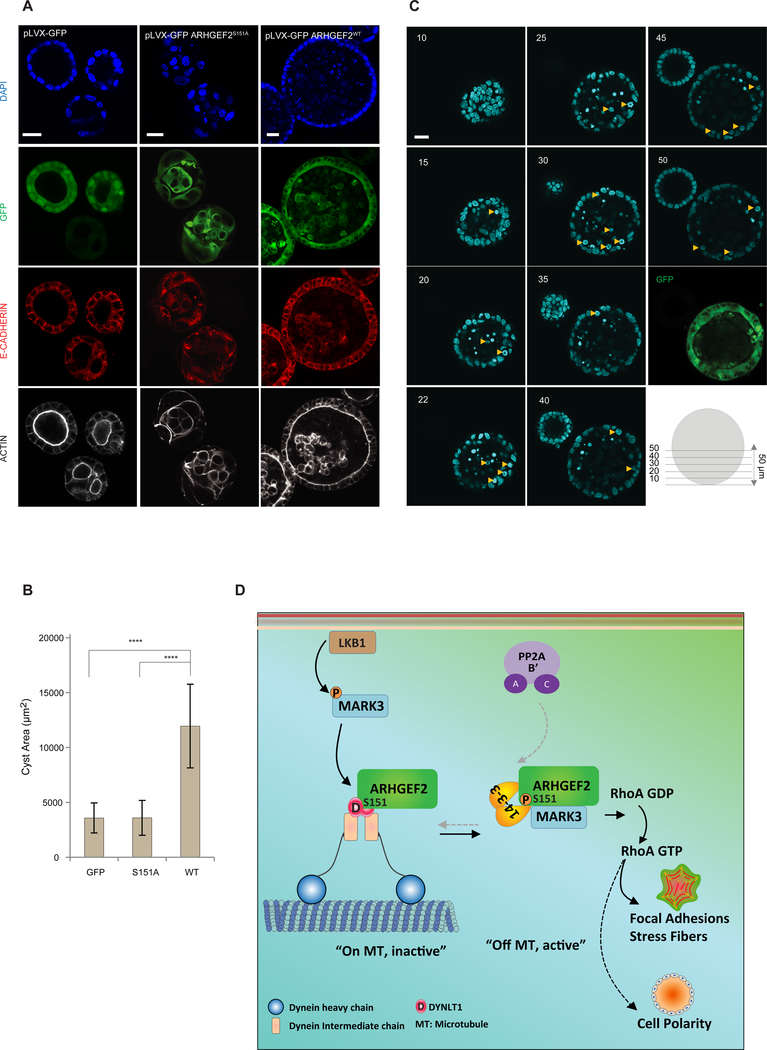
To determine the role of Ser151 phosphorylation in cell polarity we grew pLVX-GFP, pLVX-GFP ARHGEF2WT and pLVX-GFP ARHGEF2S151A expressing cells in 3D culture. Cells expressing GFP alone initiated lumen formation at day four, while those expressing pLVX-ARHGEF2WT had already established a well-defined lumen. In distinction, the pLVX-ARHGEF2S151A cells failed to initiate the formation of proper 3D structures at day four (Figure S8A). After eight days of growth in matrigel, the pLVX-GFP control cells developed a spherical structure with a hollow lumen. Expression of pLVX-GFP ARHGEF2WT led to larger spheroid structures with maintained polarity but with decreased luminal clearance of cells (Figure 8A, B). In contrast, expression of pLVX-GFP ARHGEF2S151A led to smaller spheroid with disorganized arrangement of cells, which were unable to form polarized structures with well-defined lumen (Figure 8A, B).
Figure 8. Phosphorylation of ARHGEF2 Ser151 is required for normal cell polarity.
(A-C) 3D culture of MDCKII cells stably expressing inducible pLVX-GFP, pLVX-GFP ARHGEF2WTand pLVX-GFP ARHGEF2S151A. In (A), GFP fluorescence was visualized and cysts were stained for E-CADHERIN, ACTIN and DAPI. In (B), average size of the cysts observed in pLVX-GFP, pLVX-GFP ARHGEF2WTand pLVX-GFP ARHGEF2S151A (n=24; 21; 24 respectively). Data are are means ± SD of three independent experiments. Statistical significance was determined by a one way ANOVA test with a Bonferroni post-test correction for multiple comparisons. ****P≤0.0001. Scale bar, 20 μm. In (C) 1 μm Z-stacks of pLVX-GFP ARHGEF2WT-cysts. Abnormal mitotic events are indicated (yellow arrows). Note that the cyst on the left has lost expression of pLVX-GFP ARHGEF2wt. Numbers represent the Z-stack step. Images are representative of four independent experiments. Scale bar, 20 μm.
(D) Schematic representation summarizing the effects of MARK3 and PP2A in the regulation of ARHGEF2 phosphorylation and its effects on RHOA activation. LKB1 activates MARK3 that in turn phosphorylates ARHGEF2 on Ser151. This creates a 14-3-3 binding site that disrupts ARHGEF2 interaction with DYNLT1 and releases it from microtubules to activate RHOA and formation of stress fibers and focal adhesions. MARK3 phosphorylation of Ser151 is required for epithelial cell polarity in three-dimensional growth. PP2A dephosphorylates Ser151 through interactions with the B’ subunits.
To better understand the dynamics of tight junction in these spheroids we first analyzed the distribution of the tight junction proteins zonula occludens 1 (ZO-1, also known as Tight junction protein 1 or TJP1) and 2 (ZO-2 or TJP2) in two-dimensional cultures. We observed differences in the distribution of ZO-1 and ZO-2 between the control pLVX-GFP cells versus the mutant pLVX-GFP ARHGEF2S151A (Figure S8B). The fine homogeneous staining of ZO-1 and ZO-2 observed along cell junctions in control cells was attenuated in cells expressing ARHGEF2S151A, particularly between adjacent cells, consistent with defective tight junction formation. On the other hand, pLVX-GFP ARHGEF2wt expressing cells showed an enhanced ZO-1 and ZO-2 signal at the junction of cells, compared to the control pLVX-GFP (Figure S8B).
In 3D cultured cells, we observed a classical distribution of ZO-1 in the lumen of pLVX-GFP cells. This signal was strongly enhanced in pLVX-GFP ARHGEF2WT cells with an atypical distribution of the ZO-1 staining at the lumen as well as the periphery of the spheroid. In contrast, the ZO-1 signal in pLVX-GFP ARHGEF2S151A cells was attenuated (Figure S8C). These observations reinforce our claim that cells expressing ARHGEF2S151A dominantly disturb normal sequence of cellular polarization required for epithelial cyst formation in 3D.
Multiple mitotic figures were evident in the polarized epithelial layer of the spheres in pLVX-GFP ARHGEF2WT cells suggestive of enhanced mitosis (Figure 8C). We observed enhanced proliferation in spheroids formed by pLVX-ARHGEF2WT cells (Ki67 staining), with some luminal cells positive for Ki67 (Figure S8D). Active Caspase 3 was detected in subsets of those luminal cells (Figure S8E), showing that these cells are in a dynamic state of proliferation and cell death. These data demonstrate that ARHGEF2 contributed to spheroid size and the maintenance of epithelial polarity, whereas, a mutant form of ARHGEF2 uncoupled from MARK3 interfered with the signalling program required to establish epithelial polarity and normal spheroid formation.
Discussion
We used a proteomic approach to identify ARHGEF2 as a substrate of MARK3, and mapped a unique MARK3 phosphosite on Ser151 of ARHGEF2, which regulates the interaction of ARHGEF2 with the dynein motor light chain DYNLT1 through the creation of a 14-3-3 binding site. We solved a crystal structure of DYNLT1 in complex with an ARHGEF2 peptide comprising the binding site, which reveals that the ARHGEF2 residues 140–155 lie in the groove formed at the interface of the two homodimeric DYNLT1 subunits. We showed that phosphorylation of the Ser151 site per se is insufficient to disrupt this complex. Rather, phosphorylation of Ser151 created a higher affinity 14-3-3 binding site which antagonized ARHGEF2 binding to DYNLT1 through direct competition for the binding site. 14-3-3 proteins frequently regulate the intracellular localization of their target proteins by the masking of subcellular targeting sequences such as nuclear localization sequences. Occluding the binding site between a dynein (DYNLT1) and its cargo protein (ARHGEF2) represents a previously unknown mechanism of regulation of protein subcellular localization by 14-3-3 proteins (45, 74, 75). We also showed that PP2A restored the dephosphorylated state of Ser151, and thus returned it to the inactive state associated with the microtubule array. Consistent with this model, previous studies have observed increased stress fiber formation upon inhibition of PP2A-family phosphatases with OA (76), which may be mediated through inhibition of ARHGEF2 dephosphorylation. Thus, phosphorylation of Ser151 served as a reversible switch leading to the displacement and activation of ARHGEF2 from the microtubule-associated dynein motor complex, and subsequent activation of RHOA, which affected three-dimensional cell growth (Figure 8D).
The release from microtubules and activation of ARHGEF2 can thus be regulated by three distinct modes. First, direct disruption of polymerized microtubules by pharmacologic agents such as Nocodazol or Colchicine release ARHGEF2 and lead to potent activation of RHOA, an effect that can be reversed by the microtubule stabilizing drug Paclitaxel (27, 77–79). Second, the G protein-coupled receptors (GPCR) ligands lysophosphatidic acid or thrombin trigger activation of ARHGEF2 through the disassembly of the ARHGEF2:dynein multi-protein complex by the concerted action of Gα and GPγ. Ga binds directly to ARHGEF2 and displaces it from DYNLT1, while Gβγ binds to DYNLT1 and disrupts its interaction with the dynein intermediate chain, resulting in the release of ARHGEF2 from microtubules (30). Third, we showed here that phosphorylation of the Ser151 regulatory site on the N-terminus of ARHGEF2 by MARK3 disrupted its binding to DYNLT1 by creating a 14-3-3 binding site, triggering its release from the microtubule network and activation.
The identification of MARK3 as a kinase which creates a 14-3-3 binding site on ARHGEF2 is consistent with a more general function of MARK family members in controlling substrate function or activity through the creation of 14-3-3 binding sites (40, 65, 72, 73). The cell cycle regulated phosphatase CDC25C is sequestered in the cytoplasm by MARK3-dependent phosphorylation and subsequent binding to 14-3-3. Similarly, phosphorylation of KSR1 by MARK3 creates a 14-3-3 binding site which impairs its ability to translocate to the plasma membrane and fully activate the MAPK pathway.
ARHGEF2 can be stabilized in an active state when 14-3-3 is bound to phosphoSer151 in the N-terminus or in an inactive state associated with polymerized microtubules when 14-3-3 is bound to phosphoSer885 in the C-terminus. Similarly, RAF1 can be locked in an active or an inactive state by 14-3-3 binding to distinct C-terminal or N-terminal sites, respectively. PP2A can also activate or inactivate RAF1 by differential dephosphorylation of these regulatory sites (80, 81).
14-3-3 proteins recognize sites containing phosphorylated serine or threonine residues in the context of peptide sequences containing RSx(pS/pT)xP (mode I), RxF/Yx(pS/pT)xP (mode II) (45, 82–84)or phosphosites at the penultimate C-terminal position (x(pS/pT)x-COOH) (82, 83, 85), as well as a variety of non-canonical motifs. We suggest that the ARHGEF2 Ser885 phosphosite contained within the sequence RRRpSLP is a high affinity 14-3-3 binding site that conforms to the mode I motif, whereas the Ser151 phosphosite contained within the sequence KSVpSTT is a lower affinity suboptimal mode I sequence (http://scansite3.mit.edu/) (86) since it lacks the Pro residue that directs the C-terminus of the peptide out of the binding cleft. A similar site that lacks proline at position +2 and has a lysine at position −3 instead of arginine was identified in the cytoplasmic tail of P2 integrin, where phosphorylation of threonine within the sequence KSApTTT creates a binding site for 14-3-3, which competes with and displaces talin (87).
Further work will be required to understand the physiologic input which controls MARK3 activation. The MARK family of kinases include four family members that regulate the cell cycle and cell polarity, and in turn these kinases are phosphorylated and activated by LKB1 and atypical protein kinase C (aPKC) (15, 63, 88). MARK3 is constitutively active in cells (89), and yet the determinants of kinase regulation are not currently known.
MARK3 is an abundantly phosphorylated protein containing at least 17 phosphorylation sites identified in vivo. Five MARK3 phosphorylation sites are followed by a proline residue, suggesting that they may be phosphorylated by proline-directed kinases, such as the mitogen-activated protein kinases, cyclin-dependent protein kinases or glycogen synthase kinase 3 (65, 90,91). Some of these sites represent 14-3-3 binding sites that may control the subcellular localization of MARK3, which is localized in the cytoplasm when phosphorylated and bound to 14-3-3, whereas a MARK3 mutant lacking all 17 known phosphorylation sites and unable to bind to 14-3-3 is enriched on the plasma membrane (65). The mechanism by which LKB1 activates MARK3 remains unknown. Both MARK2 and MARK3 maintain some activity in LKB1−/− mouse embryonic fibroblasts and LKB1-null HELA cells (63), suggesting that other kinases may activate MARK kinases and contribute to the basal Ser151 phosphorylation observed in LKB1-deficient A549 cells. Nevertheless, we have provided genetic and biochemical evidence that LKB1 stimulated ARHGEF2 Ser151 phosphorylation by MARK3. Further work is required to determine how the state of cell polarization affects the distribution and stoichiometry of MARK3 phosphorylation sites.
ARHGEF2 can interact with all four members of the MARK family and form a complex with the Helicobacter pylori bacterial oncoprotein CagA (92). MARK2 phosphorylates ARHGEF2 on Ser885 and Ser959 leading to the inhibition of the GEF activity with suppression of RHOA activation and stress fiber formation (36). Microtubule-associated proteins MAP2, MAP4 and MAPT/TAU are well known substrates of MARK kinases (93–95), with an emerging role for MARK4 in the phosphorylation of MAPT/TAU in Alzheimer Disease (96). The identification of ARHGEF2 and other microtubule associated proteins including CLASPs, which are involved in planar cell polarity in Arabidopsis thaliana by mediating the orientation of cell division planes in roots (97), as MARK3 substrates suggest that MARK3 is part of a protein network that regulates microtubule function, and highlights the importance of the microtubule network in cell polarity. To our knowledge, ARHGEF2 is the only well characterized MARK3 substrate linked with cell polarity in which the mutation of a single phosphosite leads to profound defects in three dimensional growth. Our results reflect the importance of phosphorylation of ARHGEF2 in Ser151 in controlling cell polarity in three-dimensional growth in mammalian cells and are consistent with previous observations in which the lack of ARHGEF2 in Xenopus lead to severe defects in neuronal tube closure due to defects in polarity linked with alterations in myosin II light chain phosphorylation and accumulation of Rab11 and actin (98). Recently it has been reported that ARHGEF2 interacts with Zonula occludens 2 (ZO2) in MDCK cells and that lack of ZO2 leads to an increase in RHOA activation mediated by ARHGEF2. The lack of ZO2 also causes misorientation of the mitotic spindle and promotes the formation of multiple lumens in cysts in three-dimensional culture, effects associated with RHOA and CDC42 activation (99).
Within this MARK3 interaction network we also identified the PP2A regulatory subunit B alpha (PPP2R2A) and the scaffold subunit A alpha (PPP2R1A) as well as several tyrosine-protein phosphatases (PTPN3, PTPN13 and PTPN14) that could influence microtubule dynamics in highly polarized epithelial cells. Since other members of the MARK family interact with ARHGEF2 we cannot exclude the possibility that they could contribute to the regulation of cell polarity through ARHGEF2 as well (92).
ARHGEF2 is a multifunctional guanine exchange factor involved in RHOA activation and MAPK pathway activation through its non-catalytic scaffold function. We have uncovered a previously unknown phospho-regulatory switch, which regulates the subcellular localization and activity of ARHGEF2. The LKB1-MARK3 pathway and PP2A dynamically control the phosphorylation state of ARHGEF2, required for the establishment of cell polarity.
Materials and Methods
Cell lines and cell culture
All cell lines were maintained in a 5% CO2 environment at 37°C and grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen), supplemented with regular or tetracycline free 10% FBS (WISENT Inc.) for the doxycycline-inducible cell lines. The following cell lines were used in this study: HEK293T (ATCC), Cos cells (D.K. Morrison, National Cancer Institute, Frederick, MD); MDCKII (S. Muthuswamy, Beth Israel Deaconess Medical Center/ Harvard Medical); 293T-Rex cells expressing Flag-tagged PP2A and PP6 subunits and Flag-tagged GFP (A.C. Gingras, Samuel Lunenfeld Research Institute, Toronto, ON). Untransfected 293 Flp-In T-REx cells (Invitrogen) were additionally supplemented with zeocin (100μg/ml) and blasticidin (5μg/ml). After transfection and selection of stable lines expressing Flag-BirA* alone or Flag-BirA*-ARHGEF2, according to the instructions of the manufacturer, the cells were maintained with hygromycin B (100μg/ml).
Transfection
Plasmid DNAs were transfected into HEK293T and 293T-Rex cells using LipoD293 (SignaGen®) according to the manufacturer’s instructions. Transfected cells were kept for 24–48 hrs in fully supplemented medium.
3D culture
An 8-well chambered coverglass system (Thermo Scientific™ Nunc™ Lab-Tek™ II) was coated evenly with 50 μl of Matrigel ® Matrix (Corning) per well and incubated at 37°C for 40 minutes to allow the matrix to solidify. MDCKII cells were trypsinized, counted and a total of 5×103 cells were resuspended in 400 ul of 3D culture media (DMEM + 10% FBS + 2% of Matrigel +/− 1 μg/ml of doxycycline) and placed on top of the solidified matrix. The cells were allowed to grow for four or eight days, with fresh 3D culture media replenishment every four days.
Live imaging
HEK293T were seeded onto micro plates 24 well, ibiTreat (Ibidi, Germany). Subconfluent cells were transfected as previously indicated, with pEGFP-C1 or pcDNA3 Cherry tagged vectors. After 24 hrs of transfection, live-cell imaging of HEK293T was performed on an inverted confocal microscope (Olympus IX81 Inverted Microscope) with a built in incubator maintained in a 5% CO2 environment at 37°C using a 60x/1.2 U PlanApo Water objective (Nikon), and FluoView software (Olympus, Tokyo, Japan).
Gene ontology, Network, motifs scan and ARHGEF2 orthologs analysis
Gene ontology (GO) analysis of the network (100, 101) was performed using all the interactors (http://geneontology.org). A further analysis, using the GO curated terms and literature reports, was done to manually classify six genes that remained unclassified (ANKRD52, ANKRD28, CEP170, CEP170B, PPP6R2 and MTUS1). The final network was built with Cytoscape_v3.1.0 (102) using shared interactions based on GeneMANIA (103)(www.genemania.org/). The motif scan was done using ScanProsite (104, 105) (http://prosite.expasy.org/scanprosite) submitting the sequences of CLASP1 (Q7Z460), CLASP1 (O75122) and ARHGEF2 (Q92974) according to their UniProt Knowledgebase (UniProtKB) accession entries. ARHGEF2 orthologs were obtained using UniProtKB (106) (http://www.uniprot.org), and reviewed (marked with an asterisk in Fig. 2E) and unreviewed orthologs were selected for the analysis. The UniProtKB sequences were aligned using PRofile ALIgNEment (PRALINE) (107,108) (http://www.ibi.vu.nl/programs/pralinewww/).
Immunoblots and co-immunoprecipitation assays
Cells were lysed in NP-40 lysis buffer as previously described (73) for analysis of the interactions with MARK3, whereas Triton X-100 lysis buffer (50 mM Hepes, 150 mM NaCl, 1% TX-100, 10% glycerol, 1mM EGTA, 1mM EDTA, protease and phosphatase inhibitors (ThermoScientific)) was used for analysis of interactions with PP2A and PP6 subunits. After the indicated treatment or transfection the cells were washed once in cold PBS and lysed for 20 min on ice. After centrifugation for 10 min at 13,000 rpm at 4°C, cleared lysates were either directly resolved by SDS-PAGE by boiling in Laemmli sample buffer 5 min and analyzed by WB or the protein complexes were incubated with antibodies overnight for endogenous IP or for 1 hour at 4°C with anti-Flag®M2 affinity gel for Flag-IP or an antibody against GFP. After the incubation period, protein-A Agarose (Roche) for GFP-IP, or protein G agarose (Bioshop) for endogenous immunoprecipitates, were added and the mixture was incubated for an additional hour at 4°C. The beads were pelleted and washed three times with cold lysis buffer. Samples were processed for SDS-PAGE by boiling in 2X Laemmli sample buffer and analyzed by WB. Quantifications were performed using the ImageJ 1.37c software and were representative of three independent experiments unless indicated otherwise.
Immune complex kinase assays
Plasmid DNAs encoding HA-tagged CLASP1, CLASP2, or ARHGEF2 were transfected into Cos cells using the Fugene reagent (Roche). 48 hrs after transfection, Cos cells were lysed under stringent conditions using radioimmunoprecipitation assay (RIPA) buffer (NP-40 lysis buffer that contains 0.5% sodium deoxycholate and 0.1% SDS). HA-tagged CLASP1, CLASP2, or ARHGEF2 proteins were immunopreciptiated from cell lysates, following which the immune complexes were washed three times with NP-40 lysis buffer and once with 30 mM Tris [pH 7.4]. The complexes were then resuspended in 40 μL of kinase buffer (30 mM Tris [pH 7.4], 1 mM DTT, 10 mM MgCl2, 5 mM MnCl2, 1 μM ATP) containing 20 μCi of [γ32P]ATP and 0.2 μg of either purified MARK3WT or a kinase-defficient MARK3kd mutant and incubated at 30oC for 30 min. The assays were terminated by the addition of gel sample buffer (250 mM Tris [pH 6.8], 50 mM DTT, 10%SDS, 30% glycerol). Samples were resolved by SDS-PAGE and the phosphoproteins were visualized by autoradiography.
Metabolic Labeling of Cells
Cos cells expressing HA-tagged ARHGEF 2 were incubated for 4–6 hr at 37°C in phosphate-free DMEM containing 2.5% dialyzed calf serum and 1 mCi of [32P] orthophosphate per ml of labeling medium. Cells were then washed twice with ice cold Tris-buffered saline (TBS) (20 mM Tris [pH 7.4]; 137 mM NaCl) and lysed in RIPA buffer. ARHGEF2 proteins were immunoprecipitated from cell lysates and washed extensively with NP-40 lysis buffer. Samples were examined by SDS-PAGE and autoradiography.
Phosphorylation Site Mapping
32P-labeled proteins were eluted from the SDS-PAGE gel matrix, precipitated using trichloroacetic acid, and digested with trypsin. An aliquot of the digested protein was adjusted to pH 2 with 20% trifluoroacetic acid and loaded onto a Waters 3.9- by 300-mm Ci8 column. Reversed-phase high-performance liquid chromatography (HPLC) was performed in an LKB chromatography system with two 2150 HPLC pumps, a 2152 LC controller, and a 2140 rapid spectral detector. When buffer salts began to elute, an increasing gradient of acetonitrile in 0.05% aqueous trifluoroacetic acid was added to the column. The stepwise gradient at a flow rate of 1 ml/min was 0 to 40% CH3CN for 60 min, 40% CH3CN for 10 min, 40 to 60% CH3CN for 10 min, and 60% CH3CN for 10 min. Fractions were collected at 1-min intervals, and 32P content was determined by measuring Cerenkov counts. HPLC fractions containing peaks of radioactivity were subjected to phosphoamino analysis and semiautomated Edman degradation in a spinning-cup sequenator as previously described (109).
Generation of stable TET-Inducible cells lines
Stable cells expressing GFP, MARK3, ARHGEF2, GFP-tagged ARHGEF2WT and GFP-tagged ARHGEF2S151A under the control of a TET-inducible promoter, were generated using the lentiviral Tet-On 3G Inducible Expression System (Clontech). Lentiviral particles were generated in HEK293T by cotransfection of the viral vectors with packaging and envelope plasmids (pPAX2 and VSV-g) using Extreme gene as previously described (26). Stable doxycycline-inducible MDCKII were established by co-transduction with two lentiviral vectors according to the instructions of the manufacturer (Clonetech): a regulator vector that stably expresses the Tet-On 3G transactivator protein and a second vector that contains the TRE3G promoter controlling the expression of un-tagged MARK3 or ARHGEF2 in a 1:1 ratio (see Figure S6 for details). The cells were selected with puromycin (10ug/ml Bioshop) and G418 (200 ug/ml Bioshop) and maintained in DMEM + 10% FBS (tetracycline free, WISENT Inc.) after antibiotic selection. Non-transduced cells were used to determine the optimal concentration of antibiotic for selection.
Antibodies and reagents
Western blotting, immunoprecipitation and immunofluorescence were performed using the following antibodies: an antibody recognizing the Pyo derived epitope tag has been reported previously (110); antibodies recognizing MARK3 and VINCULIN (immunofluorescence) from Milipore; antibodies against Flag (clone M2, Sigma) and HA (Covance); for detection of endogenous ARHGEF2 we used a mouse monoclonal antibody, clone 3C5, designed using N-terminal human ARHGEF2 peptides and produced by hybridoma as previously described (26) and an antibody recognizing a region within amino acids 656–1000 of Human ARHGEF2 (Abcam, ab155785); for endogenous immunoprecipitation of ARHGEF2 we used the ARHGEF2 specific antibody A301–929A (Bethyl); antibodies recognizing 14-3-3 (all isoforms), Myc (9E10), alpha tubulin and ZO-2 (immunofluorescence) from Santa Cruz Biotechnology (Santa Cruz, CA); antibodies recognizing ERK1/2, pARHGEF2 Ser886, pARHGEF2 Ser151 (custom produced), AMPKα, pAMPKα Thr172, LKB1, GAPDH and Cleaved Caspase 3 (immunofluorescence) from Cell Signaling; Alexa Fluor® 647 Phalloidin from Thermo Fisher Scientific; antibody specific for ZO-1 from Life Technologies; antibody specific for Ki67 from Abcam (immunofluorescence). The following reagents were used: AICAR (Santa Cruz, CA) and Okadaic Acid (Bioshop). For siRNA treatments Lipofectamine® RNAiMAX (Thermofisher Scientific) was used for the transfection of 10 nM of pre-designed siRNA control or MARK3 s230619 and s69595 (Silencer® Select, Thermofisher Scientific).
Expression constructs
Full-length, truncated and mutated human ARHGEF2 constructs (NM_001162384.1) were cloned into the 3XFlag pCMV.10 vector (Sigma), pEGFP-C1 (Clontech) or pcDNA™3.1/myc-His(–)B (Invitrogen) and verified by sequence. Bovine DYNLT1 (accession no. NM_174620) and murine 14-3-3ζ (YWHAZ) were cloned into 3XFlag pCMV.10 or pGEX-4T-3 vectors for recombinant protein generation as described previously (28). pcDNA3 constructs encoding pyo-KSRl N′424 (residues 1–424) and full length pcDNA3-Pyo-MARK3WT and -MARK3kd (Kinase deficient) have been described previously (40,110). Additionally, full length ARHGEF2 and MARK3 were cloned into the TREG vector (Clontech) and into pcDNA3 mCherry vectors. Point mutations were introduced by site-directed mutagenesis (QuickChange; Stratagene) and deletions were introduced by designing forward and reverse primers flanking the region to be deleted (Q5® Site-Directed Mutagenesis Kit; New England Biolabs, Inc.).
For structural analyses of DYNLTL1, a pGEX-4T-1 expression vector containing full-length murine DYNLT1 cDNA was a gift from Dr. Mingjie Zhang (Hong Kong University of Science and Technology, Hong Kong, China). Murine ARHGEF2 (AF177032) constructs (136–161, 136–164, and a variant with a cysteine substitution, Cys136–161) were generated by annealing DNA oligomers with BamHI- and EcoRI-compatible overhangs, which were ligated into a digested pGEX4T-1 vector to encode thrombin-cleavable glutathione S-transferase (GST)-fused fragments. A chimeric construct of ARHGEF2 residues 136–164 fused to the C-terminus of DYNLT1 through a linker of sequence GLEGGSGGSG was generated by amplifying the ARHGEF2 region with primers encoding the additional linker sequence flanked by Xhol restriction sites. The TGA stop codon of DYNLT1 in the pGEX-4T-1 expression vector was mutated to GGA using Quikchange and the vector and PCR product were each digested by Xhol, ligated together and screened for correct orientation by Sanger sequencing. The linker sequence was subsequently shortened to a single glycine using Quikchange.
Protein expression for structural analysis
E. coli BL21(DE3) CodonPlus-RIL (Agilent) cells were transformed with the DYNLT1, DYNLT1/ARHGEF2 chimera, or murine ARHGEF2 constructs and grown overnight in Lysogeny Broth (LB) medium supplemented with 50pg/mL chloramphenicol and 100pg/mL ampicillin at 37°C while shaking. These cells were pelleted by centrifugation and transferred into fresh LB for expression of unlabeled protein, or M9 media supplemented with 1g/L 15N-ammonium chloride and/or 2g/L 13C-glucose to produce isotopically labelled (13C and/or 15N) DYNLT1 or DYNLT1/ARHGEF2 chimera. Protein expression was induced by adding 250μM isopropyl-β-D-1-thiogalactopyranoside (IPTG) when the optical density (OD600) of the culture reached ~0.8. The temperature was decreased to 15°C and protein expression continued for approximately 19 hours. The cells were harvested by centrifugation and the cell pellets were flash frozen and stored at −70°C until purification.
Protein and peptide purification
Frozen BL21 cell pellets expressing DYNLT1 or DYNLT1/ARHGEF2 chimera were thawed on ice and resuspended in lysis buffer (50mM Tris-HCl pH 8.0, 300mM NaCl, 0.5mM EDTA pH 8.0, 0.1% NP-40, and 2mM DTT). Cells were lysed by sonication and the cell debris was removed by centrifugation. Glutathione Sepharose 4B resin (GE Healthcare Life Sciences) was mixed with the supernatant and incubated at 4°C for 1 hour. The resin was extensively washed with a high salt buffer 1 (50mM Tris-HCl pH 8.0, 300mM NaCl, and 2mM DTT) followed by a lower salt buffer 2 (50mM Tris-HCl pH 8.0, 150mM NaCl, and 2mM DTT). The resin was resuspended, and DYNLT1 was cleaved from the GST tag by thrombin (10units/mg of bound protein) for 48 hours at 4°C, then further purified on a Superdex 75 (S75) 26/60 size exclusion column (GE Healthcare Life Sciences) run with 50mM Tris-HCl pH 7.0, 400mM KCl, and 2mM DTT. Fractions containing pure DYNLT1 or DYNLT1/ARHGEF2 were collected and concentrated for NMR and crystallographic studies. ARHGEF2 peptides were purified in a similar manner using the following buffers: lysis buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 10% glycerol, 10mM β-mercaptoethanol, and 0.1% NP-40); wash buffer 1 (50mM Tris-HCl pH 7.5, 500mM NaCl, 10% glycerol, and 10mM β-mercaptoethanol); wash buffer 2 (50mM Tris-HCl pH 7.5, 150mM NaCl, 10% glycerol, and 10mM β-mercaptoethanol) and size-exclusion chromatography was performed using a Superdex Peptide 10/300 GL column (GE Healthcare Life Sciences) run with (50mM Tris-HCl pH 7.0, 100mM KCl, and 0.2mM TCEP). Peptide concentrations were calculated by the absorbance at 214nm using a JASCO spectrophotometer and purity was determined by silver staining (Invitrogen) Tris-Tricine SDS-PAGE gels.
NMR spectroscopy
Two dimensional 1H-15N heteronuclear single quantum coherence (HSQC) spectra of 0.2 to 0.3 mM samples were acquired on Bruker Avance III 600 MHz and Bruker Avance II 800 MHz spectrometers at 25°C in buffer consisting of 50mM Tris-HCl pH 7.0, 100mM KCl, 2mM DTT, 10% D2O. Three dimensional spectra (HNCA, HNCOCA, CBCACONH, HNCACB, HNCO, HNCACO and nuclear Overhauser effect spectroscopy (NOESY)) for backbone assignments of the DYNLT1/ARHGEF2 chimera were acquired on 0.3mM samples at 800MHz in 50mM phosphate buffer pH 6.7, 400mM KCl, 2mM DTT, 10% D2O at 35°C. NMR data was processed with NMRDraw and NMRPipe (111), while spectra were analyzed with NMRView (82).
Paramagnetic relaxation enhancement (PRE)
ARHGEF2 peptides (residues 137–158) were synthesized with an additional cysteine at either the N- or C-terminus (Lifetein), and a tryptophan at the opposite terminus to enable quantification by UV280 absorption. Peptides were conjugated to a maleimide-linked EDTA tag (N-[2-maleimidoethyl]ethylenediamine-N,N,N”,N”-tetraacetic acid) (Toronto Research Chemicals) overnight at room temperature in the presence of 0.2mM TCEP, then either paramagnetic Mn+2 (MnCl2) or diamagnetic Ca+2 (CaCl2) was added. After 2 hours, excess metal ions and unconjugated tag were removed by dialysis against 25mM HEPES pH 7, 100mM KCl, 0.2mM TCEP overnight at room temperature. Spectral perturbations of 15N-labelled DYNLT1 induced by addition of the ARHGEF2 peptides bound to Ca+2 or Mn+2 ions were compared to localize the N-terminus of the peptide through additional peak broadening associated with the conjugated paramagnetic Mn+2.
Crystallography
The DYNLT1/ARHGEF2 chimera with the optimized single glycine linker was crystallized at a protein concentration of 400μM in a hanging drop by the vapour diffusion method using a buffer comprised of 2.33M ammonium sulphate ((NH4)2SO4), 0.25mM sodium malonate pH 7 (C3H2O4Na2), and 7.2mM CaCl2. Crystals were allowed to grow for a few weeks, then harvested, soaked in 1M sodium malonate pH 7 as a cryoprotectant (112), and flash frozen in liquid nitrogen. Diffraction and data collection were performed at the synchrotron x-ray source (Advanced Photon Source (APS) at Argonne National Laboratory), and phasing was solved by molecular replacement using the Drosophila DYNLT1 structure (PBD ID 1YGT). Flexible portions of the structure were deleted while non-conserved residues were modified to alanines.
Microscale thermophoresis (MST) and Fluorescence polarization (FP)
Four 16-mer ARHGEF2 peptides comprising amino acids 876–891 with and without phosphorylation of Ser885 (RRPLDPRRR[(p)Ser885]LPAGDA[Lys(FITC)]) and amino acids 142–157 with and without phosphorylation of Ser151 (SSLSLAKSV[(p)Ser151]TTNIAG[Lys(FITC)]) were synthetized with a C-terminal FITC fluorescent tag (Biomatik Corp.). 14-3-3 protein was expressed as a recombinant GST fusion protein and purified by glutathione Sepharose followed by size-exclusion chromatography. The affinity between these ARHGEF2 peptides and 14-3-3 protein was measured by MST (Monolith NT.115, NanoTemper,) by preparing a series of standard MST glass capillaries containing 100 nM of each fluorescent peptide and increasing concentrations of GST-14-3-3 (serial dilutions from 195 μM to 2 nM) in 20 mM HEPES pH 7.4, 150 mM NaCl, 1 mM DTT, 0.05 % Tween20. Data was analyzed using the NanoTemper analysis software package. To investigate the effect of Ser151 phosphorylation on binding of DYNLT1, a series of samples containing 10 nM ARHGEF2 peptides (142–157 with and without Ser151 phosphorylation) and increasing concentrations of GST-DYNLT1 (serial dilutions from 1.15 mM to 70 nM) were analyzed by fluorescence polarization (FP) using a Molecular Devices SpectraMax M5. Although Kd values could not be determined because the binding curves did not reach saturation, phosphorylation had no appreciable effect on the binding curve.
BioID
Proximity-dependent biotinylation (BioID)(37), was performed as previously described (32). In this assay, the promiscuous mutant biotin ligase BirA* (BirAR118G) is expressed as a fusion with the protein of interest such that it will biotinylate interacting as well as proximal proteins, which can then be identified by affinity purification and mass spectrometry. Full length human ARHGEF2 (NM_001162384.1) was amplified by PCR using specific primers (Forward: 5’-GGC GCG CCA CCA TGT CTC GGA TCG AAT CCC TC-3’; Reverse: 5’-AGT TAG GCG GCC GCT TAG CTC TCG GAG GCT ACA GC-3’) and cloned into the pcDNA5 FRT/TO FlagBirA* expression vector using AscI and NotI restriction enzymes. Using the Flp-In system (Invitrogen), 293 T-REx Flp-In cells stably expressing FlagBirA* alone or the FlagBirA*-ARHGEF2 fusion were generated. After selection (DMEM + 10% FBS + 200 μg/ml Hygromycin B), sub-confluent cells were incubated overnight in complete media supplemented with 1 μg/ml tetracycline (Sigma) and 50 μm biotin (BioShop). Cells were collected and pelleted (2000 rpm, 3 min), the pellets were washed twice with PBS, dried and snap frozen.
Biotin-streptavidin affinity purification for MS
The cell pellets were resuspended in 10 mL of lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, 1:500 protease inhibitor cocktail (Sigma-Aldrich), 1:1000 TurboNuclease (Accelagen) and incubated on an end-over-end rotator at 4°C for 1 hour, briefly sonicated to disrupt any visible aggregates, then centrifuged at 16,000g for 30 min at 4°C. Supernatant was transferred to a fresh 15 mL conical tube. 30 μL of packed, pre-equilibrated Streptavidin Sepharose beads (GE) were added and the mixture was incubated for 3 hours at 4°C with end-over-end rotation. Beads were pelleted by centrifugation at 2000 rpm for 2 min and transferred with 1mL of lysis buffer to a fresh Eppendorf tube. Beads were washed once with 1 mL lysis buffer and twice with 1 mL of 50 mM ammonium bicarbonate (pH=8.3). Beads were transferred in ammonium bicarbonate to a fresh centrifuge tube, and washed two more times with 1 mL ammonium bicarbonate buffer. Tryptic digestion was performed by incubating the beads with 1 μg MS-grade tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Promega) dissolved in 200 μL of 50 mM ammonium bicarbonate (pH 8.3) overnight at 37°C. The following morning, an additional 0.5 μg trypsin was added and the beads were incubated two additional hours at 37°C. Beads were pelleted by centrifugation at 2000g for 2 min, and the supernatant was transferred to a fresh Eppendorf tube. Beads were washed twice with 150 pL of 50 mM ammonium bicarbonate, and these washes were pooled with the first eluate. The sample was lyophilized and resuspended in buffer A (0.1% formic acid). Each mass spectrometry analysis was performed on an Aliquot of 1/5th of this sample.
Mass Spectrometry
Mass spectrometry was performed as previously described (34). Briefly, analytical columns (75-um inner diameter) and pre-columns (150-um inner diameter) were made in-house from fused silica capillary tubing (InnovaQuartz) and packed with 100 Á C18-coated silica particles (Magic, Michrom Bioresources). Peptides were subjected to liquid chromatography (LC)-electrospray ionization-tandem mass spectrometry, using a 120 min reversed-phase (100% water-100% acetonitrile, 0.1% formic acid) buffer gradient running at 250 nl/min on a Proxeon EASY-nLC pump in-line with a hybrid LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). A parent ion scan was performed in the Orbitrap using a resolving power of 60,000, then the most intense peaks (up to twenty) were selected for MS/MS (minimum ion count of 1000 for activation), using standard collision-induced dissociation fragmentation. Fragment ions were detected in the LTQ. A dynamic exclusion protocol was activated such that MS/MS of the same m/z (within a range of 15 ppm; exclusion list size equal to 500) detected twice within 15 s were excluded from analysis for 30 s. For protein identification, Thermo .RAW files were converted to the .mzXML format using Proteowizard (113), then searched using X!Tandem (114) against the human (Human RefSeq Version 45) database. X!Tandem search parameters were: 15ppm parent mass error; 0.4 Da fragment mass error; complete modifications, none; cysteine modifications, none; potential modifications, +16@M and W, +32@M and W, +42@N-terminus, +1@N and Q. Data were analyzed using the trans-proteomic pipeline (TPP) (115), (116) via the ProHits software suite (117). Proteins identified with a Protein Prophet cut-off of 0.9 and at least 2 unique peptides were analyzed with the SAINT express algorithm (v3.3) (118). Fourteen control runs (consisting of fourteen Flag-BirA*only) were collapsed to the three highest spectral counts for each prey. A bayesian false discovery rate (BFDR) of 0.02 or higher, corresponding to a significance analysis of interactome (SAINT) score >0.79, was used to identify the bona fide interactors (119). The interactions are from two independent biological replicates of Flag-BirA*-ARHGEF2 (Denoted as A and B).
Isolation of MARK3 complexes and Mass Spectrometry Analysis
The experiment was performed as previously described using cycling Cos cells expressing pyo-tagged MARK3WT (60).
Immunofluorescence
For immunofluorescence (IF) imaging, subconfluent MDCKII cells were seeded on sterile Nunc™ Lab-Tek™ II Chambered Coverglass, and treated the following day with 10–500 ng/ml doxycycline (Bio Basic Canada Inc) or DMSO only as indicated for 18–24hrs. The cells were washed once with phosphate-buffered saline (PBS 1X) and visualized directly for live imaging, or fixed with 4% paraformaldehyde at room temperature for 15 min or 20 min for 3D culture cells. After fixation the cells were washed three times with 1X PBS and permeabilized with 0.1% Triton X-100 for 10–15 min at room temperature (RT), washed three times with 1X PBS and blocked with 1X PBS, 1% BSA,0.1% Triton X-100, 5% normal goat serum for 1 hr at room temperature. After blocking, the coverglass chambers were incubated at 4 °C overnight with antibody specific for VINCULIN (Millipore), diluted 1:300 in 1X PBS, 1% BSA, 0.1% Triton X-100 or at room temperature for 2 hrs for 3D culture with an antibody against E-CADHERIN (BD Biosciences), 1:1000. For the ZO-1, ZO-2, Cleaved Caspase 3 (diluted 1:100) and Ki67 (1:200) staining, the spheroids were permeabilized with 0.5% Triton X-100 for 15 min at RT and the incubation with the primary antibody was done overnight at RT. After incubation with primary antibodies the chambers were washed three times in 1X PBS and incubated with antibodies against Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor® 488, 568 or 647 conjugate secondary antibody (Invitrogen) and/or 1:50 Alexa Fluor® 647 Phalloidin for actin staining, for 1 hr at room temperature. After incubation with the secondary antibody, the chambers were washed three times in 1X PBS and, if required, the nuclei were stained with 4’, 6-diamidino-2-phenylindole (DAPI, Molecular Probes, Invitrogen) for 10 min followed by two additional washes with 1X PBS. Confocal imaging was performed with an Olympus IX81 inverted microscope using a 60x/1.4 PlanApo Oil or a 60x/1.20 Water (Nikon) objectives.
Statistical Analyses
Values are expressed as means and error bars represent means ± SD (Standard deviation). Statistical comparisons were made with Prism, version 6.0e (GraphPad) and specific tests are indicated for each figure where a statistical analysis was performed. All the tests were two-sided. A P value of P <0.05 was considered statistically significant. Bonferroni adjustment was conducted to adjust for multiple comparisons for most of the tests. For the purpose of this study the symbol meaning is *P≤0.05; **P≤0.01; ***P≤0.001;****P≤0.0001.
Supplementary Material
Data file 1. ARHGEF2 interactors reported in this study –Raw Data.
Fig. S1. ARHGEF2 interaction network.
Fig. S2. Full alignment of ARHGEF2 orthologs in vertebrates.
Fig. S3. Quantitative analysis of the effect of MARK3 phosphorylation of ARHGEF2 on its interactions with DYNLT1 and 14-3-3.
Fig. S4. Structural characterization of the DYNLT1-ARHGEF2 interaction.
Fig. S5. MARK3 but not PP6 regulates the phosphorylation of ARHGEF2 Ser151.
Fig. S6. Generation and validation of inducible cells.
Fig. S7. MARK3 affects the biological activity of ARHGEF2.
Fig. S8. Phosphorylation of ARHGEF2 Ser151 is required for normal cell polarity
Table S1. ARHGEF2 interactors reported in this study-Summary.
Table S2. GO enrichment analysis of the ARHGEF2 network.
Table S3. Functional Annotation of ARHGEF2 interactors.
Table S4. Analysis of MARK3/C-TAK1 complexes.
Table S5: Data collection and refinement statistics for DYNLT1:ARHGEF2 chimera.
Acknowledgements
We gratefully acknowledge Josée Normand for technical assistance, Genevieve Gasmi-Seabrook for assistance with NMR spectroscopy and Jonathan St-Germain for assistance up-loading the MS data. The authors thank Susan Keezer (Cell Signaling Technology) for the generation of phospho-specific antibodies used in this study and Trevor Moraes for the MST instrument access. Finally, we would like to thank Melany Wagner, Fabrice Sircoulomb and Oliver Kent for helpful discussions during development of the study.
Funding: This work was supported by the Canadian Cancer Society Research Institute (CCSRI: 704107) for R.R. Work in the Ikura lab was supported by a grant from Cancer Research Society (CRS 14014). R.R holds the Amgen Chair for Cancer Research at Princess Margaret Cancer Centre. M.I. holds the Canada Research Chair in Cancer Structural Biology. The NMR facility at UHN is supported by Canada Foundation for Innovation.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Data and materials availability: The mass spectrometry proteomics data have been deposited to MassIVE (Mass Spectrometry Interactive Virtual Environment, a member of the ProteomeXchange Consortium) (https://massive.ucsd.edu/ProteoSAFe/static/massive.isp) with the reference numbers: ARHGEF2: MassIVE MSV000081222 and CTAK (MARK3): MassIVE MSV000081223. The crystal structure was deposited at the Protein Data Bank (PDB; http://www.rcsb.org/pdb/). PDB ID [5WI4]. Plasmids require a material transfer agreement from University Health Network Technology Development and Commercialization Office.
References and Notes
- 1.Guo S, Kemphues KJ, par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611–620 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Tomancak P, Piano F, Riechmann V, Gunsalus KC, Kemphues KJ, Ephrussi A, A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat Cell Biol 2, 458–460 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Bright NJ, Thornton C, Carling D, The regulation and function of mammalian AMPK-related kinases. Acta Physiol (Oxf) 196, 15–26 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Kemphues KJ, Priess JR, Morton DG, Cheng NS, Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311–320 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Cox DN, Lu B, Sun TQ, Williams LT, Jan YN, Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr Biol 11, 75–87 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E, MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297–308 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Trinczek B, Brajenovic M, Ebneth A, Drewes G, MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J Biol Chem 279, 5915–5923 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Chen YM, Wang QJ, Hu HS, Yu PC, Zhu J, Drewes G, Piwnica-Worms H, Luo ZG, Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc Natl Acad Sci U S A 103, 8534–8539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biernat J, Wu YZ, Timm T, Zheng-Fischhofer Q, Mandelkow E, Meijer L, Mandelkow EM, Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol Biol Cell 13, 4013–4028 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazaro-Dieguez F, Cohen D, Fernandez D, Hodgson L, van Ijzendoorn SC, Musch A, Par1b links lumen polarity with LGN-NuMA positioning for distinct epithelial cell division phenotypes. J Cell Biol 203, 251–264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducharme NA, Hales CM, Lapierre LA, Ham AJ, Oztan A, Apodaca G, Goldenring JR, MARK2/EMK1/Par-1Balpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin-Darby canine kidney cells. Mol Biol Cell 17, 3625–3637 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennerz JK, Hurov JB, White LS, Lewandowski KT, Prior JL, Planer GJ, Gereau R. W. t., Piwnica-Worms D, Schmidt RE, Piwnica-Worms H, Loss of Par-1a/MARK3/C-TAK1 kinase leads to reduced adiposity, resistance to hepatic steatosis, and defective gluconeogenesis. Mol Cell Biol 30, 5043–5056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun C, Tian L, Nie J, Zhang H, Han X, Shi Y, Inactivation of MARK4, an AMP-activated protein kinase (AMPK)-related kinase, leads to insulin hypersensitivity and resistance to diet-induced obesity. J Biol Chem 287, 38305–38315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurov JB, Huang M, White LS, Lennerz J, Choi CS, Cho YR, Kim HJ, Prior JL, Piwnica-Worms D, Cantley LC, Kim JK, Shulman GI, Piwnica-Worms H, Loss of the Par-1b/MARK2 polarity kinase leads to increased metabolic rate, decreased adiposity, and insulin hypersensitivity in vivo. Proc Natl Acad Sci U S A 104, 5680–5685 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurov JB, Watkins JL, Piwnica-Worms H, Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol 14, 736–741 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Akchurin O, Du Z, Ramkellawan N, Dalal V, Han SH, Pullman J, Musch A, Susztak K, Reidy KJ, Partitioning-Defective 1a/b Depletion Impairs Glomerular and Proximal Tubule Development. J Am Soc Nephrol 27, 3725–3737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saadat I, Higashi H, Obuse C, Umeda M, Murata-Kamiya N, Saito Y, Lu H, Ohnishi N, Azuma T, Suzuki A, Ohno S, Hatakeyama M, Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 447, 330–333 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Howard J, Hyman AA, Microtubule polymerases and depolymerases. Curr Opin Cell Biol 19, 31–35 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Bowne-Anderson H, Hibbel A, Howard J, Regulation of Microtubule Growth and Catastrophe: Unifying Theory and Experiment. Trends Cell Biol 25, 769–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teng J, Takei Y, Harada A, Nakata T, Chen J, Hirokawa N, Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J Cell Biol 155, 65–76 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Zhou X, Mao TC, Shi XH, Fan DL, Zhang YM, Effect of microtubule-associated protein-4 on epidermal cell migration under different oxygen concentrations. J Dermatol 43, 674–681 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Jiang YY, Shang L, Shi ZZ, Zhang TT, Ma S, Lu CC, Zhang Y, Hao JJ, Shi C, Shi F, Xu X, Cai Y, Jia XM, Zhan QM, Wang MR, Microtubule-associated protein 4 is an important regulator of cell invasion/migration and a potential therapeutic target in esophageal squamous cell carcinoma. Oncogene 35, 4846–4856 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Benais-Pont G, Punn A, Flores-Maldonado C, Eckert J, Raposo G, Fleming TP, Cereijido M, Balda MS, Matter K, Identification of a tight junction-associated guanine nucleotide exchange factor that activates Rho and regulates paracellular permeability. J Cell Biol 160, 729–740 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakiashvili E, Speight P, Waheed F, Seth R, Lodyga M, Tanimura S, Kohno M, Rotstein OD, Kapus A, Szaszi K, GEF-H1 mediates tumor necrosis factor-alpha-induced Rho activation and myosin phosphorylation: role in the regulation of tubular paracellular permeability. J Biol Chem 284, 11454–11466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birukova AA, Fu P, Xing J, Yakubov B, Cokic I, Birukov KG, Mechanotransduction by GEF-H1 as a novel mechanism of ventilator-induced vascular endothelial permeability. Am J Physiol Lung Cell Mol Physiol 298, L837–848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullis J, Meiri D, Sandi MJ, Radulovich N, Kent OA, Medrano M, Mokady D, Normand J, Larose J, Marcotte R, Marshall CB, Ikura M, Ketela T, Moffat J, Neel BG, Gingras AC, Tsao MS, Rottapel R, The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell 25, 181–195 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Meiri D, Marshall CB, Greeve MA, Kim B, Balan M, Suarez F, Bakal C, Wu C, Larose J, Fine N, Ikura M, Rottapel R, Mechanistic insight into the microtubule and actin cytoskeleton coupling through dynein-dependent RhoGEF inhibition. Mol Cell 45, 642–655 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Meiri D, Greeve MA, Brunet A, Finan D, Wells CD, LaRose J, Rottapel R, Modulation of Rho guanine exchange factor Lfc activity by protein kinase A-mediated phosphorylation. Mol Cell Biol 29, 5963–5973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zenke FT, Krendel M, DerMardirossian C, King CC, Bohl BP, Bokoch GM, p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. J Biol Chem 279, 18392–18400 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Meiri D, Marshall CB, Mokady D, LaRose J, Mullin M, Gingras AC, Ikura M, Rottapel R, Mechanistic insight into GPCR-mediated activation of the microtubule-associated RhoA exchange factor GEF-H1. Nat Commun 5, 4857 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Roux KJ, Kim DI, Raida M, Burke B, A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, Archinti M, Cheung SW, Pinchev D, Lawo S, Raught B, Bazett-Jones DP, Luders J, Pelletier L, CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol 23, 1360–1366 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T, PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci 118, 1861–1872 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, Bashkurov M, Hasegan M, Knight JD, Lin ZY, Schueler M, Hildebrandt F, Moffat J, Gingras AC, Raught B, Pelletier L, A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 163, 1484–1499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimura Y, Miki H, Dynamic regulation of GEF-H1 localization at microtubules by Par1b/MARK2. Biochem Biophys Res Commun 408, 322–328 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Yamahashi Y, Saito Y, Murata-Kamiya N, Hatakeyama M, Polarity-regulating kinase partitioning-defective 1b (PAR1b) phosphorylates guanine nucleotide exchange factor H1 (GEF-H1) to regulate RhoA-dependent actin cytoskeletal reorganization. J Biol Chem 286, 4457644584 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, Wrana JL, Choi H, Gingras AC, Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal 6, rs15 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Bhandari D, Zhang J, Menon S, Lord C, Chen S, Helm JR, Thorsen K, Corbett KD, Hay JC, Ferro-Novick S, Sit4p/PP6 regulates ER-to-Golgi traffic by controlling the dephosphorylation of COPII coat subunits. Mol Biol Cell 24, 2727–2738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogg S, Gabrielli B, Piwnica-Worms H, Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J Biol Chem 269, 30461–30469 (1994). [PubMed] [Google Scholar]
- 40.Muller J, Ritt DA, Copeland TD, Morrison DK, Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. EMBO J 22, 4431–4442 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim J, Zhou M, Veenstra TD, Morrison DK, The CNK1 scaffold binds cytohesins and promotes insulin pathway signaling. Genes Dev 24, 1496–1506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dale S, Wilson WA, Edelman AM, Hardie DG, Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett 361, 191–195 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE, A rapid method for determining protein kinase phosphorylation specificity. Nat Methods 1, 27–29 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Riou P, Kjaer S, Garg R, Purkiss A, George R, Cain RJ, Bineva G, Reymond N, McColl B, Thompson AJ, O’Reilly N, McDonald NQ, Parker PJ, Ridley AJ, 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell 153, 640–653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obsil T, Obsilova V, Structural basis of 14-3-3 protein functions. Semin Cell Dev Biol 22, 663–672 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Marshall CB, Meiri D, Smith MJ, Mazhab-Jafari MT, Gasmi-Seabrook GM, Rottapel R, Stambolic V, Ikura M, Probing the GTPase cycle with real-time NMR: GAP and GEF activities in cell extracts. Methods 57, 473–485 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi N, Freund SM, Chatellier J, Zahn R, Fersht AR, NMR analysis of the binding of a rhodanese peptide to a minichaperone in solution. J Mol Biol 292, 181–190 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Chan AW, Hutchinson EG, Harris D, Thornton JM, Identification, classification, and analysis of beta-bulges in proteins. Protein Sci 2, 1574–1590 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craveur P, Joseph AP, Rebehmed J, de Brevern AG, beta-Bulges: extensive structural analyses of beta-sheets irregularities. Protein Sci 22, 1366–1378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang JZ, Yeh TY, Bollati F, Conde C, Canavosio F, Caceres A, Sung CH, The dynein light chain Tctex-1 has a dynein-independent role in actin remodeling during neurite outgrowth. Dev Cell 9, 75–86 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC, Hendrickson WA, Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc Natl Acad Sci U S A 104, 10028–10033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR 3rd, Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res 7, 1346–1351 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP, A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 105, 10762–10767 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK, Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal 2, ra46 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R, Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A 103, 5391–5396 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaffer BE, Levin RS, Hertz NT, Maures TJ, Schoof ML, Hollstein PE, Benayoun BA, Banko MR, Shaw RJ, Shokat KM, Brunet A, Identification of AMPK Phosphorylation Sites Reveals a Network of Proteins Involved in Cell Invasion and Facilitates Large-Scale Substrate Prediction. Cell Metab 22, 907–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright PE, Dyson HJ, Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merino-Gracia J, Zamora-Carreras H, Bruix M, Rodriguez-Crespo I, Molecular Basis for the Protein Recognition Specificity of the Dynein Light Chain DYNLT1/Tctex1: CHARACTERIZATION OF THE INTERACTION WITH ACTIVIN RECEPTOR IIB. J Biol Chem 291, 20962–20975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balan M, Thesis (M Sc ), University of Toronto (Canada: ), 2013., (2013). [Google Scholar]
- 60.Ory S, Zhou M, Conrads TP, Veenstra TD, Morrison DK, Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr Biol 13, 1356–1364 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Sontag JM, Nunbhakdi-Craig V, White CL 3rd, Halpain S, Sontag E, The protein phosphatase PP2A/Balpha binds to the microtubule-associated proteins Tau and MAP2 at a motif also recognized by the kinase Fyn: implications for tauopathies. J Biol Chem 287, 1498414993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watkins GR, Wang N, Mazalouskas MD, Gomez RJ, Guthrie CR, Kraemer BC, Schweiger S, Spiller BW, Wadzinski BE, Monoubiquitination promotes calpain cleavage of the protein phosphatase 2A (PP2A) regulatory subunit alpha4, altering PP2A stability and microtubule-associated protein phosphorylation. J Biol Chem 287, 24207–24215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR, LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 23, 833–843 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang JW, Imai Y, Lu B, Activation of PAR-1 kinase and stimulation of tau phosphorylation by diverse signals require the tumor suppressor protein LKB1. J Neurosci 27, 574–581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goransson O, Deak M, Wullschleger S, Morrice NA, Prescott AR, Alessi DR, Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation. J Cell Sci 119, 40594070 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Muller O, Back W, Zimmer M, Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet 18, 38–43 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA, A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 391, 184–187 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Martin SG, St Johnston D, A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature 421, 379–384 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Watts JL, Morton DG, Bestman J, Kemphues KJ, The C elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development 127, 1467–1475 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC, Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457–466 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, Shaw RJ, LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 23, 143–158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ 3rd, Wu Z, Stephenson MT, Piwnica-Worms H, C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ 9, 197–208 (1998). [PubMed] [Google Scholar]
- 73.Muller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK, C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell 8, 983–993 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Dougherty MK, Morrison DK, Unlocking the code of 14-3-3. J Cell Sci 117, 1875–1884 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Pahuja K. Bajaj, Wang J, Blagoveshchenskaya A, Lim L, Madhusudhan MS, Mayinger P, Schekman R, Phosphoregulatory protein 14-3-3 facilitates SAC1 transport from the endoplasmic reticulum. Proc Natl Acad Sci U S A 112, E3199–3206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lontay B, Kiss A, Gergely P, Hartshorne DJ, Erdodi F, Okadaic acid induces phosphorylation and translocation of myosin phosphatase target subunit 1 influencing myosin phosphorylation, stress fiber assembly and cell migration in HepG2 cells. Cell Signal 17, 1265–1275 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Chang YC, Nalbant P, Birkenfeld J, Chang ZF, Bokoch GM, GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol Biol Cell 19, 2147–2153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fine N, Dimitriou ID, Rullo J, Sandi MJ, Petri B, Haitsma J, Ibrahim H, La Rose J, Glogauer M, Kubes P, Cybulsky M, Rottapel R, GEF-H1 is necessary for neutrophil shear stress-induced migration during inflammation. J Cell Biol 215, 107–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krendel M, Zenke FT, Bokoch GM, Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat Cell Biol 4, 294–301 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Lavoie H, Therrien M, Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol 16, 281–298 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W, Raf family kinases: old dogs have learned new tricks. Genes Cancer 2, 232–260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C, Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J 427, 69–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith AJ, Daut J, Schwappach B, Membrane proteins as 14-3-3 clients in functional regulation and intracellular transport. Physiology (Bethesda) 26, 181–191 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC, The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 (1997). [DOI] [PubMed] [Google Scholar]
- 85.Rajan S, Preisig-Muller R, Wischmeyer E, Nehring R, Hanley PJ, Renigunta V, Musset B, Schlichthorl G, Derst C, Karschin A, Daut J, Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3. J Physiol 545, 13–26 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Obenauer JC, Cantley LC, Yaffe MB, Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31, 3635–3641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takala H, Nurminen E, Nurmi SM, Aatonen M, Strandin T, Takatalo M, Kiema T, Gahmberg CG, Ylanne J, Fagerholm SC, Beta2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood 112, 1853–1862 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, Miwa Y, Ohno S, aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr Biol 14, 1425–1435 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Dequiedt F, Martin M, Von Blume J, Vertommen D, Lecomte E, Mari N, Heinen MF, Bachmann M, Twizere JC, Huang MC, Rider MH, Piwnica-Worms H, Seufferlein T, Kettmann R, New role for hPar-1 kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases. Mol Cell Biol 26, 7086–7102 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pelech SL, Networking with proline-directed protein kinases implicated in tau phosphorylation. Neurobiol Aging 16, 247–256; discussion 257–261 (1995). [DOI] [PubMed] [Google Scholar]
- 91.Ubersax JA, Ferrell JE Jr., Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 8, 530–541 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Saito Y, Murata-Kamiya N, Hirayama T, Ohba Y, Hatakeyama M, Conversion of Helicobacter pylori CagA from senescence inducer to oncogenic driver through polarity-dependent regulation of p21. J Exp Med 207, 2157–2174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Illenberger S, Drewes G, Trinczek B, Biernat J, Meyer HE, Olmsted JB, Mandelkow EM, Mandelkow E, Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J Biol Chem 271, 10834–10843 (1996). [DOI] [PubMed] [Google Scholar]
- 94.Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE, Mandelkow EM, Mandelkow E, Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem 270, 7679–7688 (1995). [DOI] [PubMed] [Google Scholar]
- 95.Lund H, Gustafsson E, Svensson A, Nilsson M, Berg M, Sunnemark D, von Euler G, MARK4 and MARK3 associate with early tau phosphorylation in Alzheimer’s disease granulovacuolar degeneration bodies. Acta Neuropathol Commun 2, 22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gu GJ, Lund H, Wu D, Blokzijl A, Classon C, von Euler G, Landegren U, Sunnemark D, Kamali-Moghaddam M, Role of individual MARK isoforms in phosphorylation of tau at Ser(2)(6)(2) in Alzheimer’s disease. Neuromolecular Med 15, 458–469 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Pietra S, Gustavsson A, Kiefer C, Kalmbach L, Horstedt P, Ikeda Y, Stepanova AN, Alonso JM, Grebe M, Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat Commun 4, 2779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Itoh K, Ossipova O, Sokol SY, GEF-H1 functions in apical constriction and cell intercalations and is essential for vertebrate neural tube closure. J Cell Sci 127, 2542–2553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raya-Sandino A, Castillo-Kauil A, Dominguez-Calderon A, Alarcon L, Flores-Benitez D, Cuellar-Perez F, Lopez-Bayghen B, Chavez-Munguia B, Vazquez-Prado J, Gonzalez-Mariscal L, Zonula occludens-2 regulates Rho proteins activity and the development of epithelial cytoarchitecture and barrier function. Biochim Biophys Acta, (2017). [DOI] [PubMed] [Google Scholar]
- 100.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gene Ontology C, Gene Ontology Consortium: going forward. Nucleic Acids Res 43, D1049–1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q, The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38, W214–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P, PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform 3, 265–274 (2002). [DOI] [PubMed] [Google Scholar]
- 105.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N, ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34, W362–365 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.UniProt C, UniProt: a hub for protein information. Nucleic Acids Res 43, D204–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bawono P, Heringa J, PRALINE: a versatile multiple sequence alignment toolkit. Methods Mol Biol 1079, 245–262 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Simossis VA, Kleinjung J, Heringa J, Homology-extended sequence alignment. Nucleic Acids Res 33, 816–824 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morrison DK, Heidecker G, Rapp UR, Copeland TD, Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem 268, 17309–17316 (1993). [PubMed] [Google Scholar]
- 110.Therrien M, Michaud NR, Rubin GM, Morrison DK, KSR modulates signal propagation within the MAPK cascade. Genes Dev 10, 2684–2695 (1996). [DOI] [PubMed] [Google Scholar]
- 111.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A, NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 112.Holyoak T, Fenn TD, Wilson MA, Moulin AG, Ringe D, Petsko GA, Malonate: a versatile cryoprotectant and stabilizing solution for salt-grown macromolecular crystals. Acta Crystallogr D Biol Crystallogr 59, 2356–2358 (2003). [DOI] [PubMed] [Google Scholar]
- 113.Kessner D, Chambers M, Burke R, Agus D, Mallick P, ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Craig R, Beavis RC, TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004). [DOI] [PubMed] [Google Scholar]
- 115.Pedrioli PG, Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol Biol 604, 213–238 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R, A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu G, Zhang J, Larsen B, Stark C, Breitkreutz A, Lin ZY, Breitkreutz BJ, Ding Y, Colwill K, Pasculescu A, Pawson T, Wrana JL, Nesvizhskii AI, Raught B, Tyers M, Gingras AC, ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol 28, 1015–1017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H, SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics 100, 37–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI, SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods 8, 70–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file 1. ARHGEF2 interactors reported in this study –Raw Data.
Fig. S1. ARHGEF2 interaction network.
Fig. S2. Full alignment of ARHGEF2 orthologs in vertebrates.
Fig. S3. Quantitative analysis of the effect of MARK3 phosphorylation of ARHGEF2 on its interactions with DYNLT1 and 14-3-3.
Fig. S4. Structural characterization of the DYNLT1-ARHGEF2 interaction.
Fig. S5. MARK3 but not PP6 regulates the phosphorylation of ARHGEF2 Ser151.
Fig. S6. Generation and validation of inducible cells.
Fig. S7. MARK3 affects the biological activity of ARHGEF2.
Fig. S8. Phosphorylation of ARHGEF2 Ser151 is required for normal cell polarity
Table S1. ARHGEF2 interactors reported in this study-Summary.
Table S2. GO enrichment analysis of the ARHGEF2 network.
Table S3. Functional Annotation of ARHGEF2 interactors.
Table S4. Analysis of MARK3/C-TAK1 complexes.
Table S5: Data collection and refinement statistics for DYNLT1:ARHGEF2 chimera.