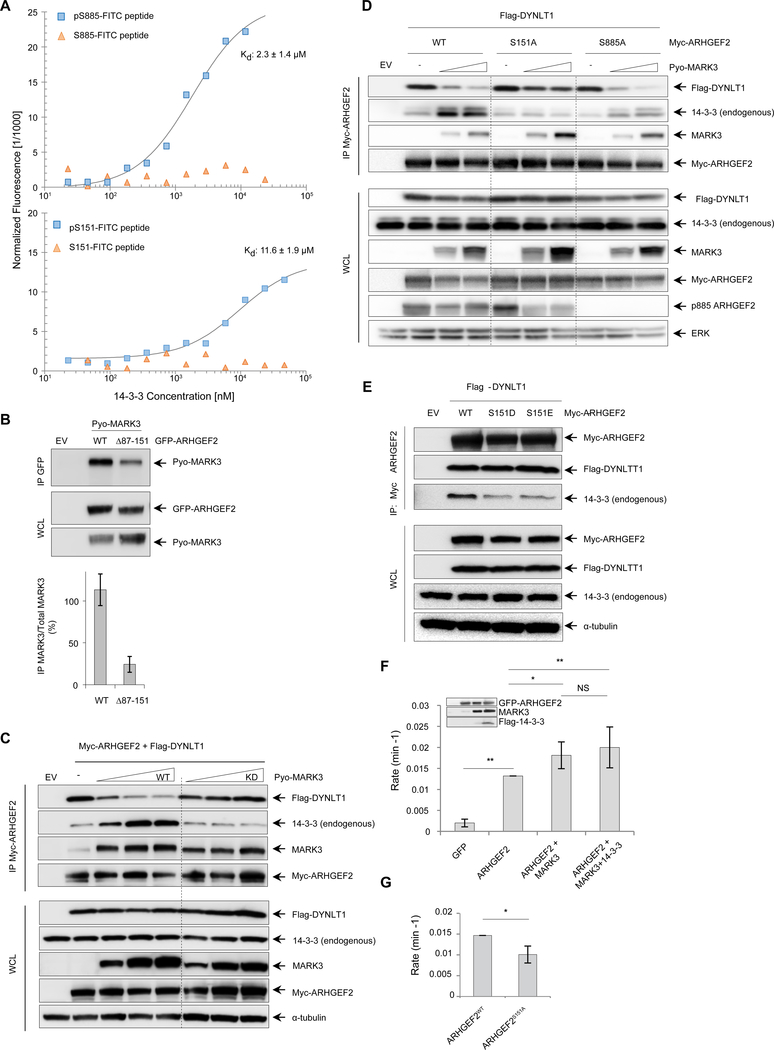
Figure 3. MARK3 perturbs the interaction between DYNLT1 and ARHGEF2 and stimulates exchange activity.
(A) Microscale thermophoresis binding assays of phosphorylated and unphosphorylated FITC-labelled ARHGEF2 peptides for Ser885 (amino acids 876–891, top panel) and Ser151 (amino acids 142–157, lower panel). The peptides were prepared at 100 nM with increasing concentrations of GST-14-3-3. Kd (dissociation constant) values were determined from the thermophoresis titration curves for the phosphorylated peptides, whereas no binding was detected for unphosphorylated peptides. Representative of three independent experiments, Kd values are average of three independent experiments ±SD.
(B) GFP-tagged wild-type ARHGEF2 and a truncated version (deletion of residues 87–151) were co-expressed with Pyo-tagged wild-type MARK3 in HEK293T cells. Protein complexes were immunoprecipitated using an antibody specific for GFP and immunoblots were probed with antibodies recognizing Pyo to detect interactions with MARK3. Antibodies specific for GFP and Pyo were used to detect protein abundance in whole cell lysates. Lower panel: quantification of the interaction normalized with total lysate. Data are means ± SD of three independent experiments.
(C) Myc-tagged ARHGEF2 was co-expressed with Flag-tagged DYNLT1 in the absence or presence of increasing amounts of wild-type (WT) or kinase-deficient (KD) MARK3. Protein complexes were immunoprecipitated using an antibody recognizing Myc antibody and analyzed by Western blot for the presence of MARK3, DYNLT1 and endogenous 14-3-3 using antibodies against MARK3, Flag and pan 14-3-3 respectively. Whole cells lysates were analyzed by Western blot and probed with the same antibodies to assess protein abundance; a-tubulin was used as a loading control. Representative of four independent experiments. See also Figure S3A, B.
(D) Myc-tagged wild-type ARHGEF2 and S151A and S885A mutants were co-expressed with Flag-tagged DYNLT1 in the absence or presence of increasing amounts of wild-type MARK3 in HEK293T cells. Protein complexes were immunoprecipitated using an antibody recognizing Myc and analyzed by Western blot for the presence of MARK3, DYNLT1 and endogenous 14-3-3 using antibodies against MARK3, Flag and 14-3-3, respectively. Whole cells lysates were analyzed with the same antibodies to assess protein abundance. Total ERK was used as a loading control. See also Figure S3C. Representative of three independent experiments.
(E) Myc-tagged wild-type and phosphomimetic mutants S151D and S151E for ARHGEF2 were co-expressed with Flag-tagged DYNLT1. Protein complexes were immunoprecipitated with an antibody recognizing Myc and analyzed by Western blot. Antibodies against Myc, Flag and 14-3-3-were used to confirm the amount of ARHGEF2 and to detect DYNLT1 and endogenous 14-3-3 in the complexes respectively. Protein abundance in whole cell lysates was analyzed using the same antibodies, and a-tubulin was used as a loading control. Representative of three independent experiments.
(F) NMR-based GEF assays were performed to measure RHOA exchange rates in the presence of cell lysates from HEK293T cells expressing GFP alone; GFP-ARHGEF2; GFP-ARHGEF2 and Pyo-MARK3; or GFP-ARHGEF2, Pyo-MARK3 and Flag-14-3-3. The amount of ARHGEF2 in exchange assays was normalized on the basis of GFP fluorescence in the lysate and protein expression concentration was detected by Western blot (inset). The rates were normalized to ARHGEF2 exchange rate. Data are means ± SD of five independent experiments. Statistical significance was determined by a Kruskal-Walis test with a Dunn’s post-test correction for multiple comparisons. *P=0.0151 (ARHGEF2 VS ARHGEF2+MARK3); **P=0.0072 (ARHGEF2 VS ARHGEF2+MARK3+14-3-3); **P=0.0079 (GFP VS ARHGEF2). NS: not significant.
(G) Nucleotide exchange rates for RHOA in the presence of cell lysates from HEK293T cells expressing GFP-ARHGEF2wt or GFP-tagged ARHGEF2S151A. The rates were normalized to ARHGEF2WT exchange rate. Data are means ± SD of four independent experiments. Statistical significance was determined by a Mann-Whitney test. *P=0.0286.