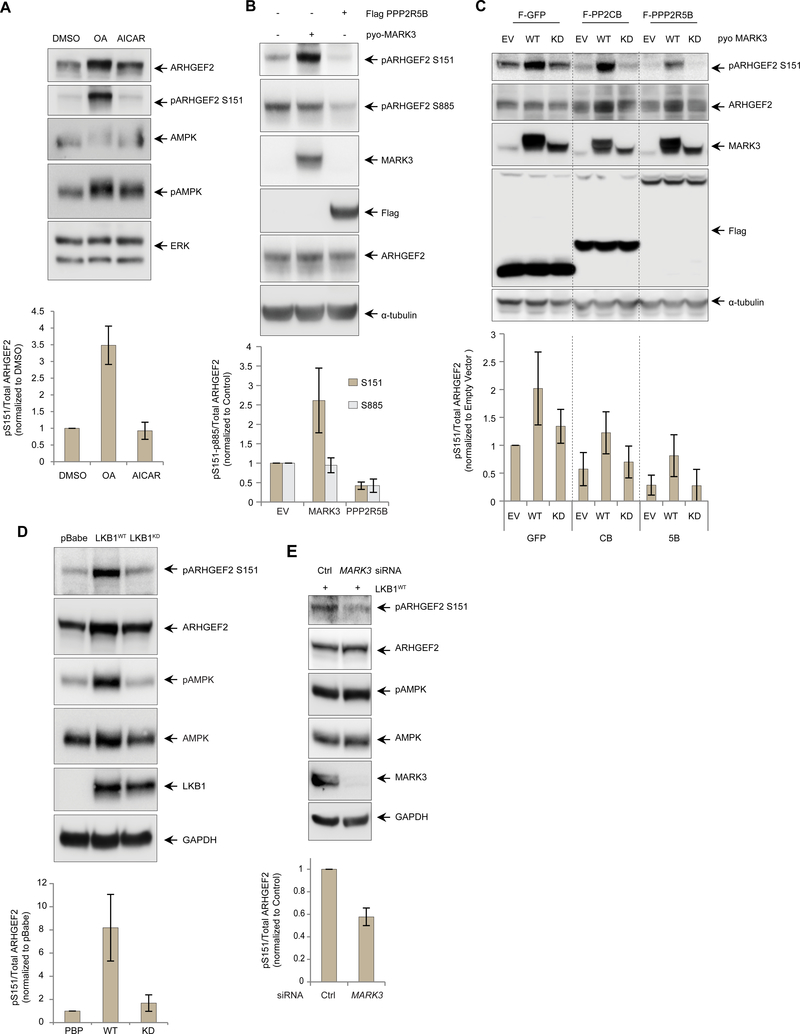
Figure 5. LKB1-MARK3 axis and PP2A regulate the phosphorylation of ARHGEF2 Ser151.
(A) Western blot of HEK293T cells treated with DMSO, PP2A inhibitor okadaic acid (OA, 50 nM for 4 hours) and AMPK activator AICAR (1mM for 6 hours). Phosphorylated ARHGEF2 Ser151 was detected using a site-specific antibody and total ERK was used as a loading control (See Figure S5 for details). Lower panel: quantification of the phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independedent experiments.
(B) Western blot of HEK293T cells overexpressing pyo-tagged MARK3WT or Flag-tagged PPP2R5B. Phosphorylated ARHGEF2 Ser151 and Ser885 were detected using site-specific antibodies and a-tubulin was used as a loading control. Lower panel: quantification of Ser151 and Ser885 phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independent experiments.
(C) Western blot of 293 T-Rex Flp-In cell lines carrying inducible expression of Flag-tagged GFP, PP2A catalytic subunit PPP2CB or regulatory B’subunit PPP2R5B. The cells were induced overnight with tetracycline 500ng/ml and also transfected with empty vector, pyo-MARK3WT or pyo-MARK3KD. Phosphorylated ARHGEF2 Ser151 was detected using a site-specific antibody and α-tubulin was used as a loading control. Lower panel: quantification of the phosphorylation normalized with total ARHGEF2, Data are means ± SD of four independent experiments.
(D) Western blot of A549 LKB1-deficient cells stably expressing empty vector (pBabe), wild-type LKB1 (LKB1WT) or kinase deficient LKB1 (LKB1KD). Phosphorylation of AMPK was used as a control substrate for LKB1 phosphorylation. Phosphorylated ARHGEF2 Ser151 and AMPK Thr172 were detected using site-specific antibodies and GAPDH was used as a loading control. Lower panel: quantification of ARHGEF2 Ser151 phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independent experiments.
(E) Western blot of A549 cells expressing LBK1WT and treated with an siRNA pool specific for MARK3 versus control for 72 hours. Phosphorylation AMPK was used as a control substrate for LKB 1 phosphorylation. GAPDH was used as a loading control. Lower panel: quantification of ARHGEF2 Ser151 phosphorylation normalized with total ARHGEF2. Data are means ± SD of three independent experiments.