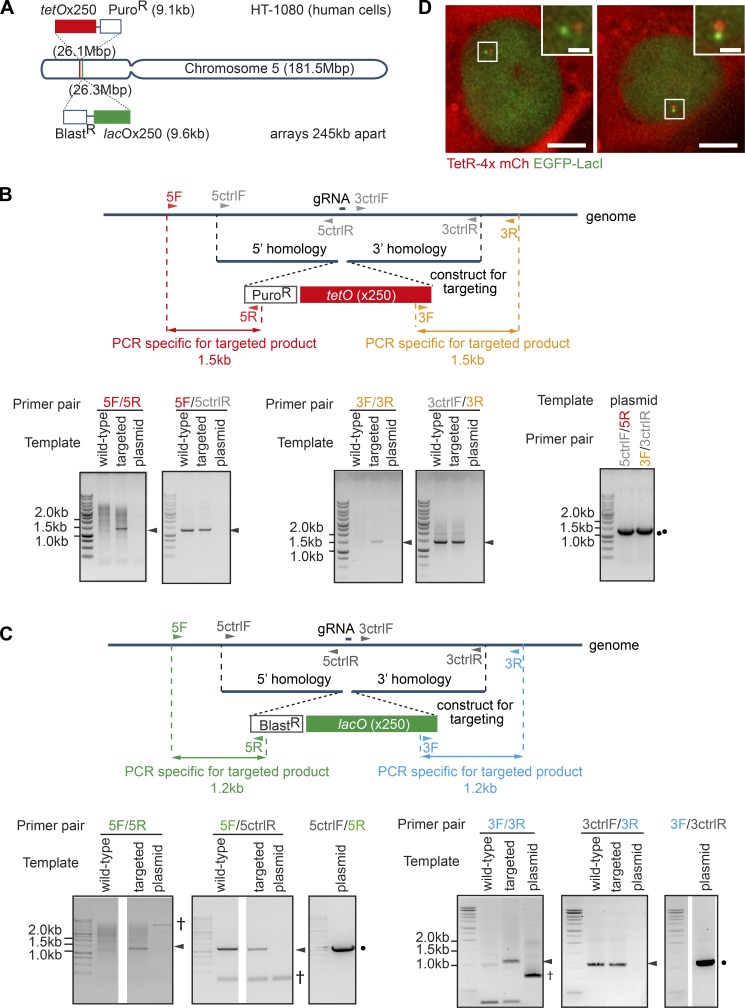
Figure 1.
Targeting tet and lac operator arrays to a selected chromosome region. (A) Diagram depicting the location of tetO and lacO introduced into human chromosome 5. (B and C) The maps (top) show the DNA construct used for targeted integration of tetO and lacO and the corresponding positions on the genome of the guide RNA (gRNA for CRISPR-Cas9) and PCR primers (e.g., 5F, 5R, etc.) for integration check. The DNA electrophoreses (bottom) show the results of PCR using template DNA from the WT genome, the genome with targeted integration (targeted), and, as control, the purified plasmid targeting construct. Arrowheads and black dots indicate PCR products expected from correct integration and the purified plasmid, respectively. Nonspecific bands are indicated by a single dagger (†). Results demonstrate correct targeted integration of tetO and lacO. (D) Visualization of these arrays by expression of TetR-4x mCherry and EGFP-LacI in HT-1080 cells (TT75). The white square boxes represent the zoomed region shown in the upper right-hand corners. Scale bars, 10 µm (main) and 1 µm (zoomed inset).