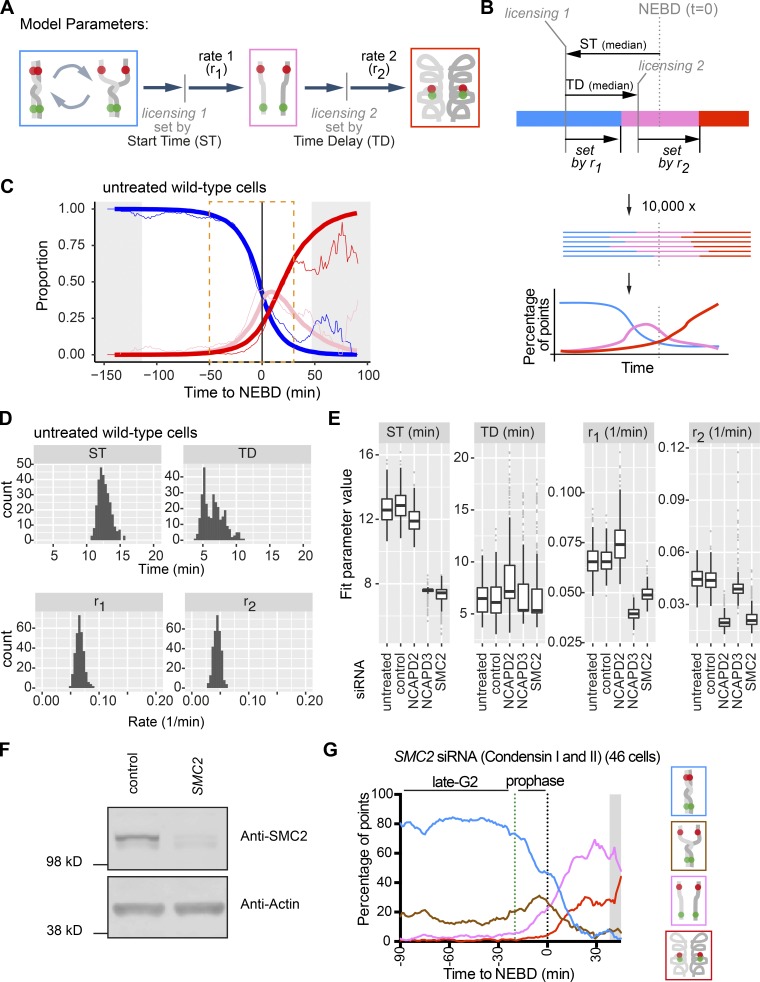
Figure 8.
Mathematical modeling of sister chromosome resolution and compaction. (A) Diagram illustrates setting of four parameters: ST, TD, r1, and r2. Also refer to B (top) for how licensing 1 and 2 were defined by ST and TD. (B) Schematic shows the procedure of modeling. Top: shows how the timing of transitions was defined by four parameters (see A). The color codes represent the chromosome configurations as in A. Note that, in a subset of cells, the transition to the pink “resolved” state happened after licensing 2; in these cells, transition to the red “compacted” state could only occur after the transition to the pink state. (C) Microscopy data (thin curves; from Fig. 2 C) and the best-fit model (thick curves) for untreated WT cells. The curves represent the proportion of each configuration of the fluorescence reporter over time. The microscopy data curves were smoothed over 5 min, while model fitting was performed on original nonsmoothed data (within the orange dotted box). The color codes are as in A and gray-shaded area is as in Fig. 3 E. (D) Distributions of best-fit parameter values were obtained from 300 bootstrapping repetitions for untreated WT cells. (E) Box-and-whisker plots of best-fit parameter values, obtained from bootstrapping (see D), for the indicated siRNA conditions. Thick horizontal lines show medians. (F) Western blotting analysis of SMC2 protein following treatment with SMC2 or control siRNA for 48 h. Actin is shown as a loading control. (G) The proportion of each configuration of the fluorescence reporter in cells depleted of SMC2. TT75 cells were treated and analyzed as in Fig. S2 I. Data from individual cells are shown in Fig. S5 A. The number of analyzed cells at each time point is between 10 and 56 (mean: 38), except for the gray-shaded areas where <10 cells were analyzed.