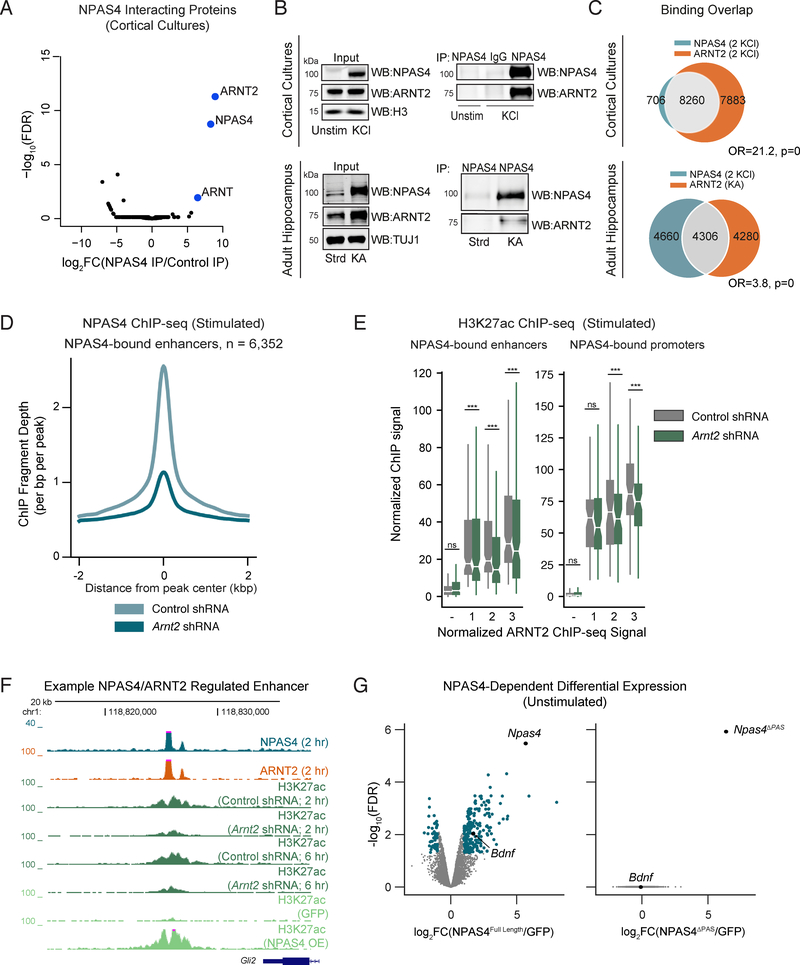
Figure 5. ARNT2 is required for NPAS4-dependent gene activation.
A.−log10(FDR) versus log2 fold change in peptides obtained by IP-MS with an anti-NPAS4 antibody in stimulated primary cortical neurons relative to peptides immunoprecipitated from control samples. Control samples include a non-specific IgG and immunoprecipitation with an anti-NPAS4 antibody from unstimulated neurons, where NPAS4 is not expressed. Blue points indicate proteins with FDR <0.1 and log2FC > 0.
B. Immunoblot analysis of anti-NPAS4 immunoprecipitates performed in cultured neurons or hippocampal tissue confirming the interaction between ARNT2 and NPAS4 both in vitro and in vivo.
C. Overlap between NPAS4 binding sites in stimulated cultured cortical neurons (2 KCl) and ARNT2 binding sites in both stimulated cultured neurons (2 KCl, top) and adult hippocampal tissue derived from mice treated with kainate (KA, bottom) to induce neuronal activity. Fisher’s exact test odds ratios (OR) and p-values for set intersections shown on figure.
D. Mean normalized NPAS4 ChIP-seq signal in control or ARNT2-depleted neurons across NPAS4-bound enhancers.
E. Normalized H3K27ac ChIP-seq signal in control shRNA (gray) and Arnt2 shRNA (green) transduced cultured neurons at NPAS4-bound enhancers (left) and promoters (right). Data plotted by quantiles of in vitro ARNT2 binding strength (3 > 1). − indicates sites not bound by ARNT2. *** p<.001, Wilcoxon signed-rank test comparing distributions of Arnt2 shRNA and control shRNA conditions by quantile.
F. UCSC genome browser ChIP-seq tracks displaying H3K27ac signal at a representative NPAS4-bound enhancer in cortical cultures stimulated with 55mM KCl. Each ChIP-seq track is labeled for the ChIP-ed factor, time point after membrane depolarization, and treatment.
G. −log10(FDR) versus log2 fold change in gene expression NPAS4Full-Length (left) or NPAS4ΔPAS (right) expressing neurons to GFP controls in unstimulated cortical neurons. NPAS4-regulated genes are indicated in blue. Npas4 and its known target gene Bdnf are labeled in both plots for reference.