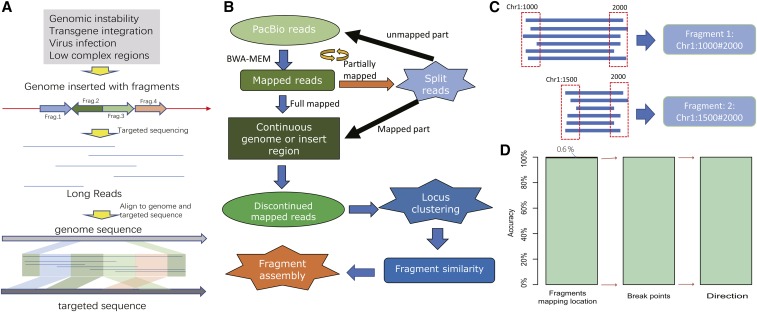
Figure 1.
The flowchart of TSD and its evaluation. (a) TSD is designed to identify the structural organization of complex SVs. In this exemplary demonstration, four pieces of exogenous DNA sequences (DNA fragments from the targeted sequences) rearrange and integrate into the host genome. TSD is used to identify their origins, rearrangement and integration location in the host genome. (b) The flowchart of TSD in dealing the PacBio reads from complex SVs. The long reads are aligned to both host genome and targeted sequence using BWA-MEM tool. If the reads are partially mapped, the unmapped fragments are cut for a new round of alignment. This can be repeated for multiple times until no unmapped fragment is longer than 200 bp. The final SV structure is determined by assembling the mapped fragments. (c) Build consensus fragments. The error-tolerating setting of BWA leads to frequent deviation from the true start and end locations. To overcome this problem, we infer the consensus start and end locations from redundant targeted sequencing reads utilizing a voting strategy. The read dash lines indicate acceptable ranges to select the reads with the same start or end location. (d) Evaluation using simulated PacBio reads. 99.4% of reads are correctly mapped to human genome; using the correctly mapped reads, 100% of simulated SVs are recovered accurately for both break point location and direction.