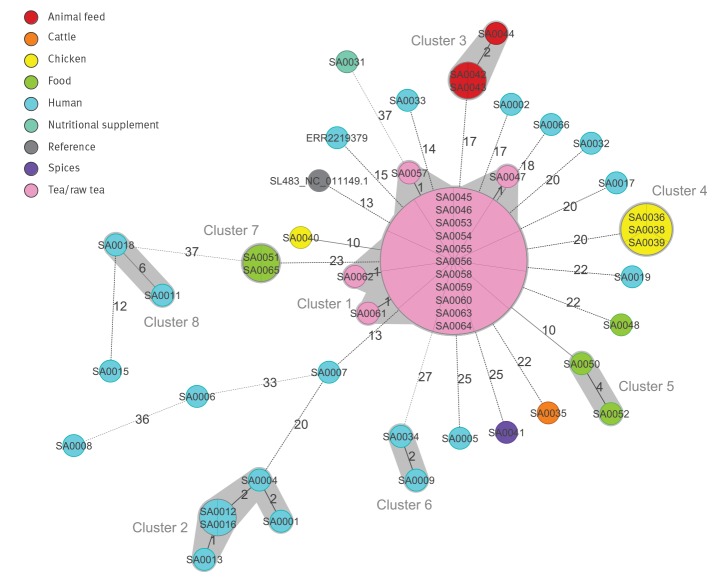
Figure 1.
Minimum spanning tree of the core genome multilocus sequence type allelic profiles of Salmonella Agona strains, including 51 Bavarian isolates, a representative isolate of an infant-milk-caused outbreak in France and the reference strain SL-483, Germany, 1993−2018 (n = 53 isolates)
cgMLST: core genome multilocus sequence typing.
Bavarian S. Agona samples have the prefix SA. The representative isolate of the infant-milk outbreak in France [23] is ERR2219379. The National Center for Biotechnology Information (NCBI) reference genome of strain SL-483 has GenBank accession number NC_011149.1.
The analysis and tree were obtained with the public Ridom-SeqSphere + -integrated S. enterica cgMLST scheme of 3,002 target loci.
On the tree, allele distances between samples are indicated. Clusters of samples with maximum seven alleles distance are shaded in grey. Samples are colour coded by their isolation source, as given in the legend.