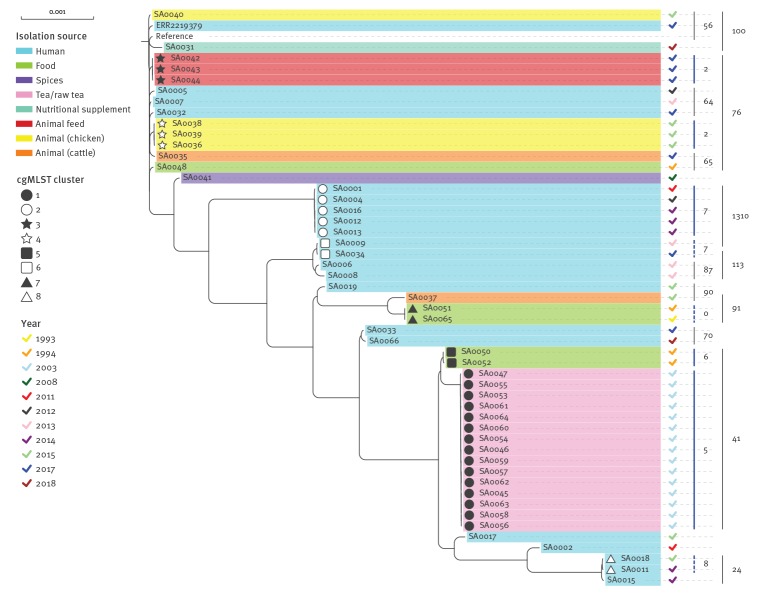
Figure 2.
Maximum likelihood tree resulting from the whole genome single nucleotide polymorphism-based phylogenetic analysis of Salmonella Agona strains including 51 Bavarian isolates, a representative isolate of an infant milk-caused outbreak in France and the reference strain SL-483, Germany, 1993−2018 (n = 53 isolates)
cgMLST: core genome multilocus sequence typing.
Bavarian S. Agona samples have the prefix SA. The representative isolate of the infant milk outbreak in France [23] is ERR2219379. The National Center for Biotechnology Information (NCBI) reference genome of strain SL-483 has GenBank accession number NC_011149.1 and figures in the tree as ‘reference’.
In the tree, samples are colour coded according to isolation source, S. Agona cgMLST cluster and collection year as given in the legend.
The general scale bar indicates 0.001 substitutions per site (597 SNPs), based on an alignment of 596,849 positions.
SNP distance bars between specific samples are indicated on the right side. Blue vertical SNP scale bars indicate epidemiologically-linked cgMLST clusters, blue vertical dashed lines indicate cgMLST clusters without epidemiological link and black vertical lines indicate exemplary distances between non clustered samples.