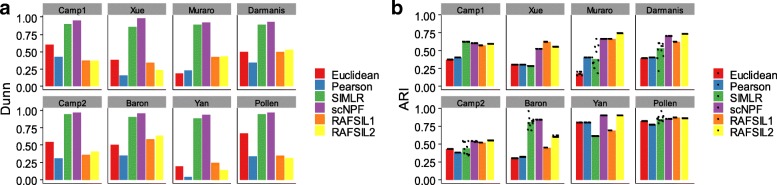
Fig. 5.
Performance comparison of the five similarity measurements on eight published scRNA-seq data sets. a The internal validation metric of Dunn was employed to measure the cell separation. b ARI is employed to measure the concordance between inferred and true cluster labels. K-means clustering is applied on the similarity matrices obtained from different methods. K-means clustering is repeated for 10 times. Each dot represents an individual K-means clustering run and each bar represents the median performance. Detailed information of the data sets is shown in Additional file 2: Table S1