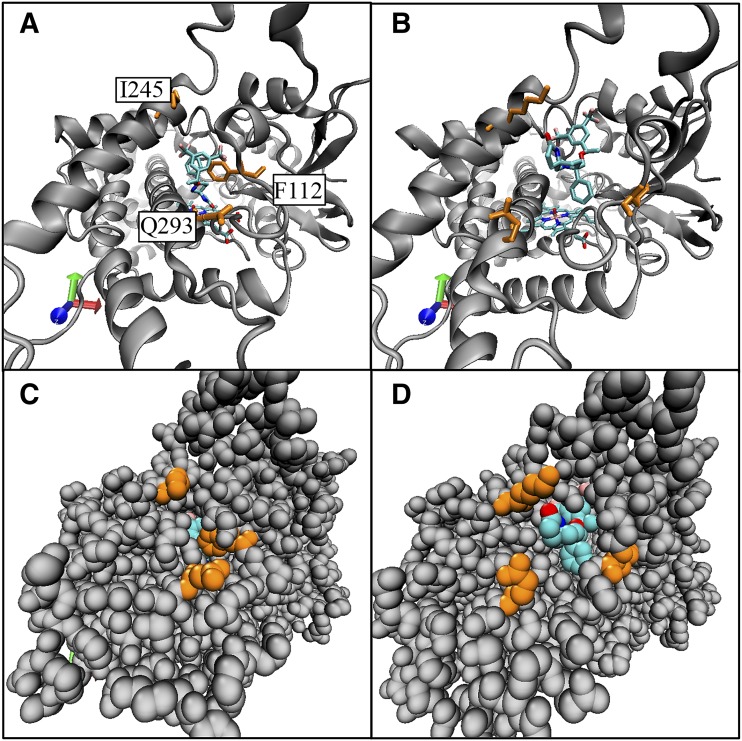
Fig. 7.
Comparison of the active site starting position to the egress position in channel 2c for rolapitant egress. Frames from molecular dynamics simulation 2 comparing the starting position of rolapitant at the active site of CYP2D6 to the egress position at the opening of channel 2c are shown. Rolapitant is shown in turquoise. Select amino acids along channel 2c are shown in orange: Phe112 on the helix B′/B-C loop, Glu293 on the N-terminal end of helix I, and Lys245 on helix G. (A) Initial position for rolapitant in the active site of CYP2D6 in a ribbon model. (B) Frame where rolapitant egresses from CYP2D6 via channel 2c. (C). Space-filling model of structure in (A); rolapitant is only slightly visible in the active site. (D). Space-filling model of structure in (B) with rolapitant emerged from channel 2c.