Fig. 7.
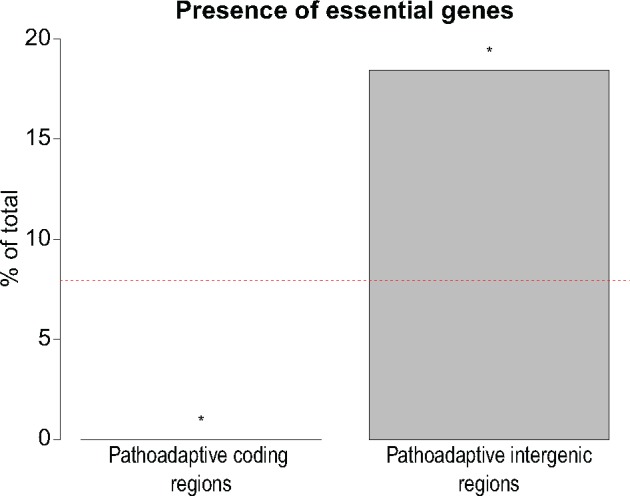
—Qualitative function of adaptive coding and intergenic mutations. A total of 52 coding regions were previously shown to be under positive selection in a selected subset of our data set comprised of 474 long-term CF-adapted isolates of P. aeruginosa (Marvig et al. 2015). In the same isolates, we identify 35 intergenic regions under positive selection for adaptive mutations (supplementary table S8, Supplementary Material online). 445 genes are essential for survival of P. aeruginosa in CF sputum environment (Turner et al. 2015). The percentage of these essential genes within 52 adaptive genes and 38 genes downstream of 35 adaptive intergenic regions is demonstrated. Asterisk denotes P < 0.05 from two-tailed Fisher’s exact test. Red dashed line indicates the percentage of CF essential genes in PAO1.
