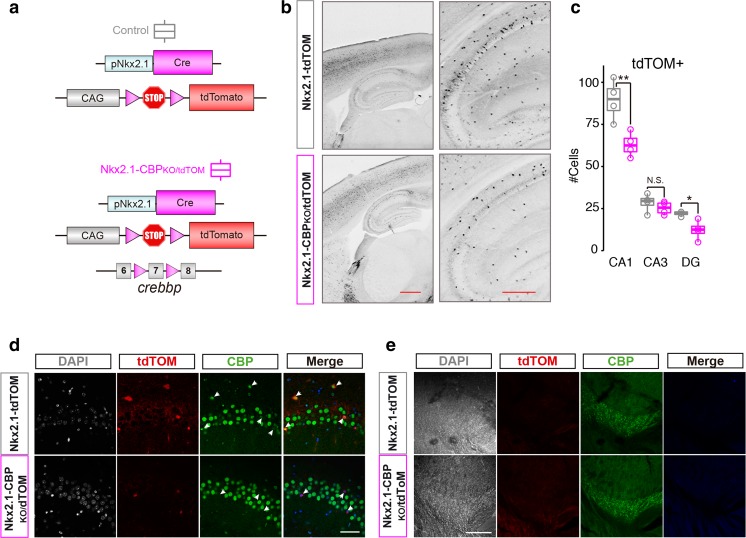
Fig. 2.
Genetic mapping demonstrates that Nkx2.1-CBPKO mice have fewer MGE-derived cells. a Scheme of the genetic mapping strategy. The Nkx2-1 promoter drives the expression of the Cre recombinase at MGE progenitors leading to the removal of a STOP cassette that otherwise prevents transcription of the red fluorescent protein tdTOM. b Representative images showing the dramatic reduction in the number of tdTOM+ cells in Nkx2.1-CBPKO/tdTOM mice. Scale bars: 500 μm. c Experimental mice have fewer tdTOM+ cells in CA1(t6 = − 3.8, p = 0.01, unpaired t test) and DG (t3,36 = − 3.30, p = 0.04, Welch t test) subdivisions of the hippocampus, but not in CA3 (t6 = − 0.97, p = 0.36, unpaired t test). d tdTOM+ cells in Nkx2.1-CBPKO/tdTOM do not express CBP. We present images of the CA1 subfield, but similar results were observed in other areas. Scale bar: 50 μm. e No differences are observed in the reticular nucleus of the thalamus. Scale bar: 200 μm. *P < 0.05; **P < 0.01