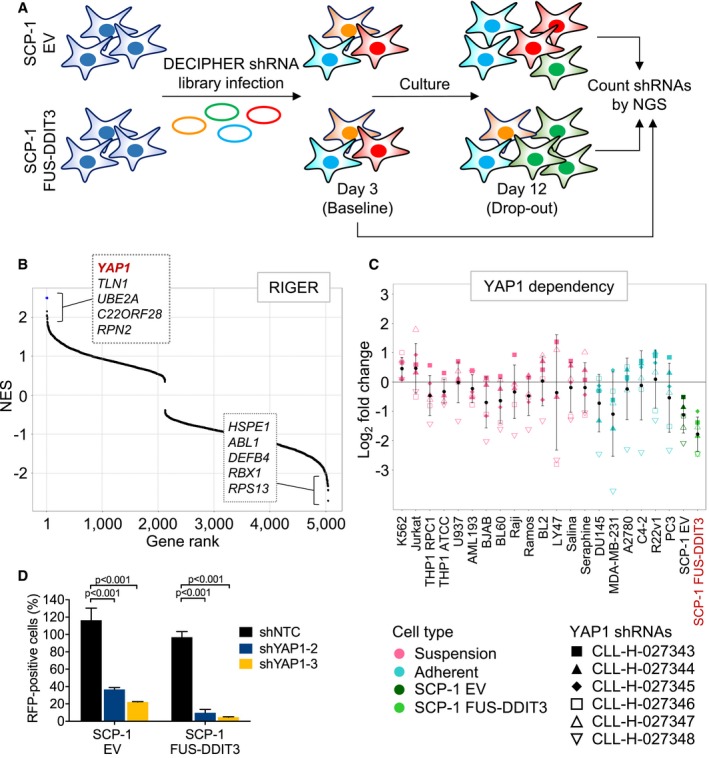
Figure 1. Identification of genes required by FUS‐DDIT3‐expressing mesenchymal stem cells.
-
ASchematic of RNAi screens. SCP‐1 cells expressing FUS‐DDIT3 or EV were transduced with Module 1 of the DECIPHER Pooled Lentiviral Human Genome‐Wide shRNA Library. Half of the cells were harvested on day 3 (baseline sample) and day 12 (drop‐out sample), respectively, and shRNA abundance was determined by next‐generation sequencing (NGS).
-
BRIGER analysis to identify genes that are preferentially essential in FUS‐DDIT3‐expressing SCP‐1 cells. EV‐transduced SCP‐1 cells and 20 FUS‐DDIT3‐negative cancer cell lines screened with the same shRNA library were used as reference set. Genes were ranked according to relative shRNA depletion, and YAP1 was identified as top FUS‐DDIT3‐specific essential gene. NES, normalized enrichment score.
-
CLFC change in YAP1 shRNA representation in 20 cancer cell lines and SCP‐1 cells transduced with FUS‐DDIT3 or EV. Black dots and error bars represent the mean ± SD of LFC scores for six independent shRNAs.
-
DCompetition assays with SCP‐1 cells transduced with RFP‐labeled NTC or YAP1 shRNAs. Flow cytometric quantification of RFP‐positive cells on day 9 relative to day 3 showed that YAP1 knockdown was preferentially toxic to FUS‐DDIT3‐expressing cultures. Bars and error bars represent the mean ± SD of two independent experiments, two‐way ANOVA.
