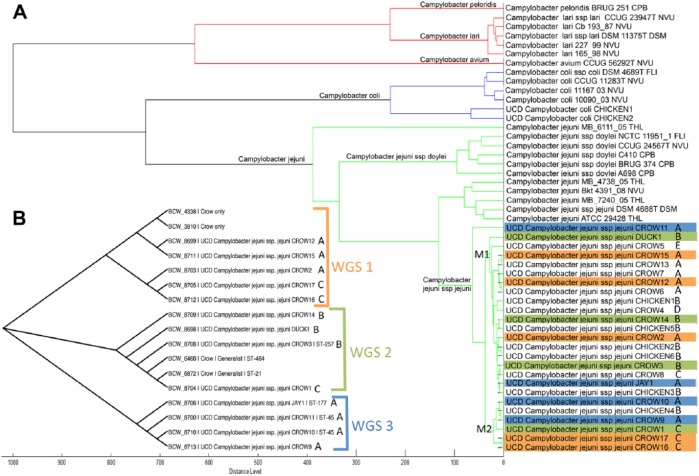
Figure 1.
A. Dendrogram showing distances and grouping of Campylobacter isolates based on protein phenotypes created using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The 2 major subclades of C. jejuni subsp. jejuni are designated M1 and M2. Isolates with similar letters (A–E) had identical 16S ribosomal (r)DNA sequences. Color-coding refers to 3 clades seen in the distance matrix tree. B. Phylogenetic relationships of 13 C. jejuni subsp. jejuni isolates determined by whole genome sequence analysis. Three clades are identified (WGS1, WGS2, WGS3) in the distance matrix tree, and isolates with identical 16S rDNA sequences are identified as above (A–E).