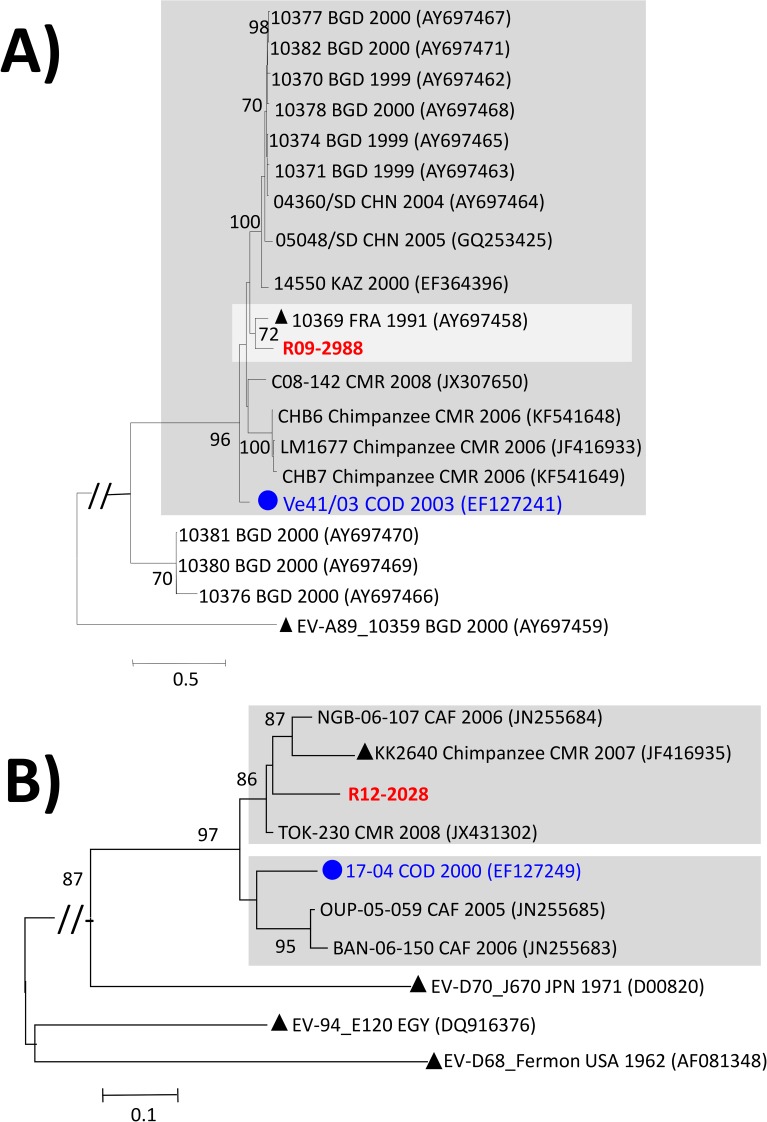
Fig 2. Phylogenetic relationships of the full-length VP1 sequences of the studied EV-A and -D.
Maximum likelihood trees were inferred from the alignments of full-length VP1 sequences of EV-A76 using the GTR+G+I model of nucleotide substitution (A) and partial VP1 sequences of EV-D111 strains (nt 1–471 according to the VP1 sequence of the EV-D111 prototype strain KK2640) using the T92+G model of nucleotide substitution (B). Studied isolates are specified in bold red while isolates previously reported in DR Congo are highlighted by circles in bold blue. The year and country of isolation of each reference isolate are indicated, when known (BGD: Bangladesh; CAF: Central African Republic; CMR: Cameroon; CHN: China; COD: Democratic Republic of the Congo; EGY: Egypt; FRA: France; JPN, Japan; KAZ: Kazakhstan; USA: United-States of America). Prototype strains are highlighted by black triangles. For clarity, bootstrap values less than 70% have been omitted and the scale bars indicate nucleotide distance as substitutions per site. Isolates belonging to specific lineages commented in the main text are gathered in grey-shaded boxes.