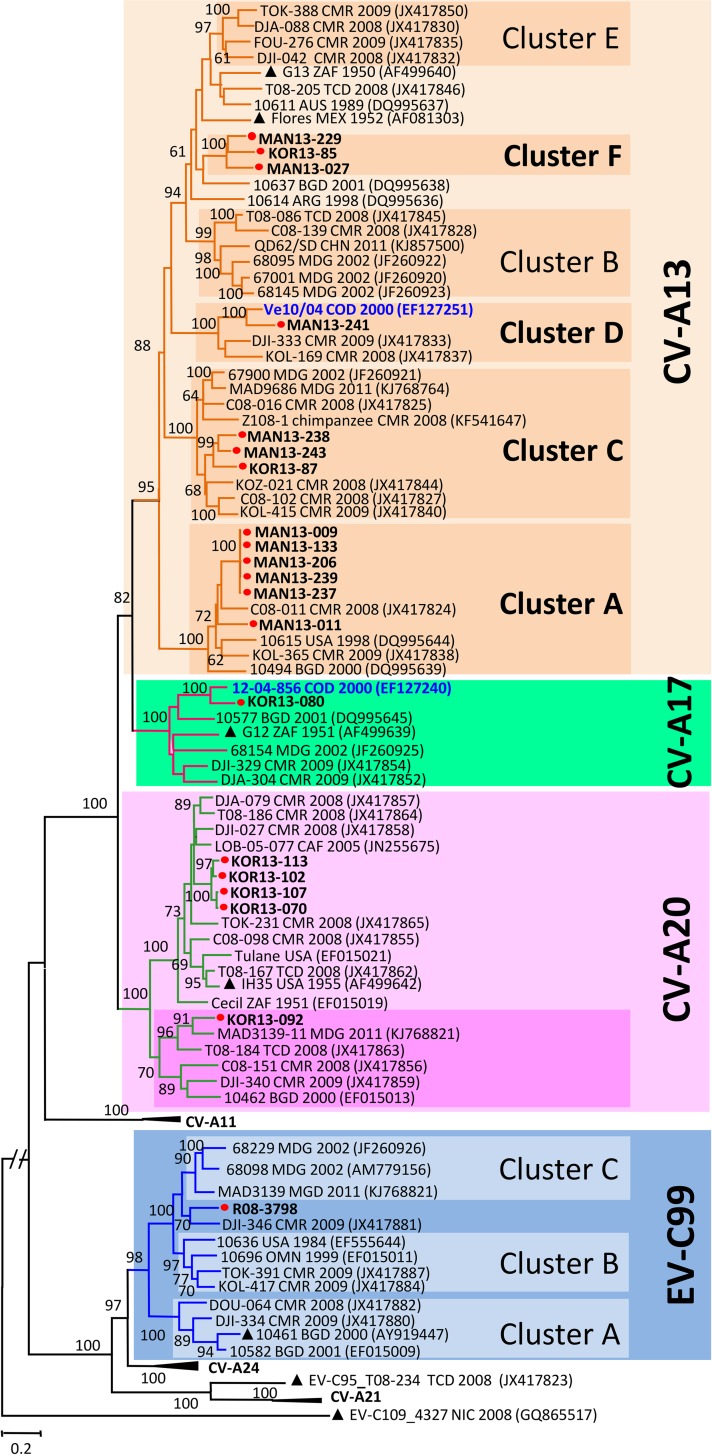
Fig 3. Phylogenetic relationships of the newly sequenced EV-C isolates from DR Congo.
The Maximum likelihood phylogram was inferred from the alignment of full-length VP1 sequences of EV-C strains using the most complex GTR+G+I model of nucleotide substitution. Previously described isolates originating from DR Congo are indicated in bold blue while isolates from this study are highlighted by red circles. Isolates from healthy children are named with a 3 letter code indicating the province of origin (MAN: Maniema and KOR: Kasai Oriental) whereas the unique EV-C99 recovered from a paralyzed child is R08-3798. The year and country of isolation of each reference isolate are indicated, when known (ARG: Argentina; AUS: Australia; BGD: Bangladesh; CAF: Central African Republic; CMR: Cameroon; CHN: China; COD: Democratic Republic of the Congo; NIC: Nicaragua; MEX: Mexico; MDG: Madagascar; OMN: Oman; TCD: Chad; USA: United-States of America; ZAF: South Africa). In addition to the sequences of prototype strains, sequences displaying highest similarities with the studied isolates were retrieved from databases using NCBI BLAST search and included as references in the analyzed dataset. Prototype strains are highlighted by triangles. For clarity, most bootstrap values less than 60 have been omitted. The scale is shown at the bottom, as substitutions per site. Isolates belonging to the virus types and lineages commented in the main text are gathered in color-shaded boxes.