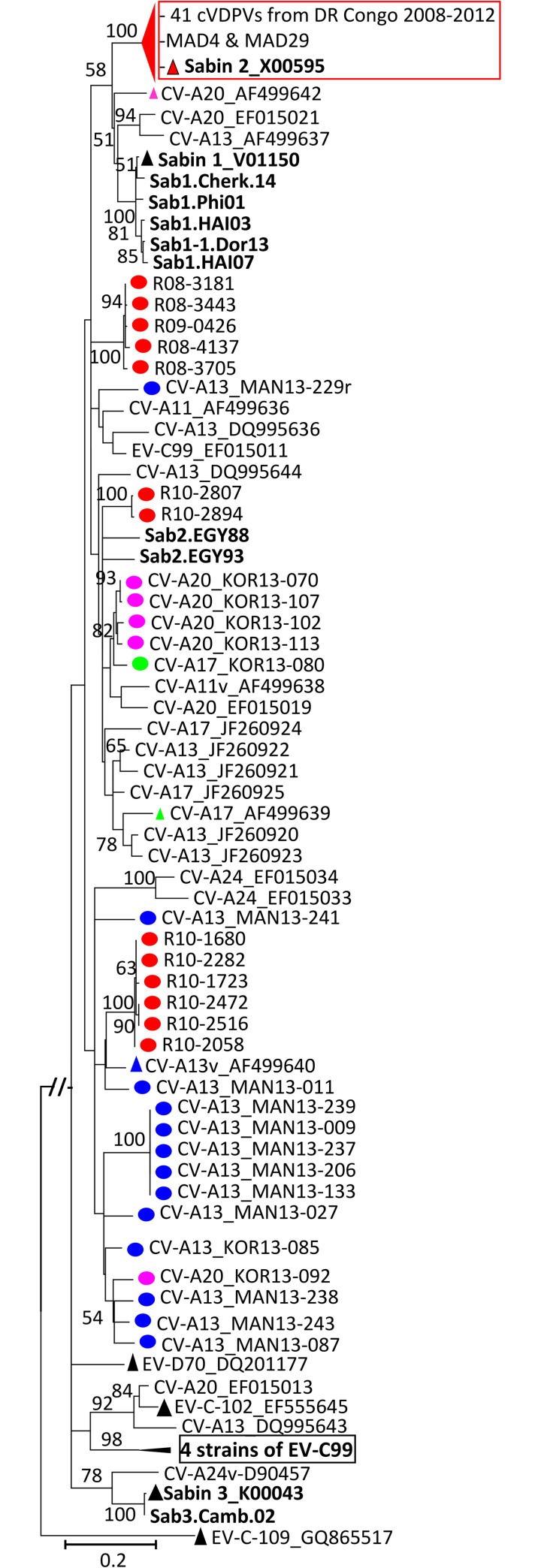
Fig 5. Phylogenetic trees depicting genetic relationships between nucleotide sequences derived from the 5’utr genomic regions of species C enteroviruses.
The maximum likelihood tree was based on the alignment of partial nucleotide sequence of the 5’UTR region of the genome (nt 183 to 573 according the complete genome of the Sabin 2). Branch lengths were calculated using the best-fit model of nucleotide substitution GTR+G+I estimated with Smart Model Selection based on the Bayesian Information Criterion. Numbers at nodes correspond to the percentage of 1,000 bootstrap replicates supporting the distal cluster. For comparison, lineages depicted on the VP1-based reference phylogeny (Fig 4A), were highlighted here despite the fact that they were not supported by well-defined and bootstrap supported groups (Fig 4A). For comparison, the cluster defined by the 41 circulating vaccine-derived polioviruses featuring Sabin 2 related sequences were collapsed. Details about isolates’ labeling and the dataset considered are the same as specified in the legend of Fig 4.