Abstract
Mitochondrial DNA is sensitive to damage by exogenous reactive oxygen sources, including traffic-related air pollution (TRAP). Given the important role for mitochondria in human disease, we hypothesized that prenatal air pollution exposure may be associated with mitochondrial dysfunction and that mitochondrial-derived peptides (MDPs) might protect against these effects. In in vitro studies, 24–hour exposure to nanoparticulate matter (nPM) increased oxidation of mtDNA, decreased mitochondrial consumption rate (OCR), and decreased mtDNAcn in SH–SY5Y cells. Addition of MDPs rescued these effects to varying degrees. Liver tissue taken from C57Bl/6 males exposed for 10 weeks to nPM had lower OCR, lower mtDNAcn and higher MDP levels, similar to in vitro studies. In newborn cord blood, MDP levels were positively associated with prenatal TRAP exposures. Moreover, DNA methylation of two distinct regions of the D-Loop in the mitochondria genome was associated with levels of several MDPs. Our in vitro and in vivo data indicate that TRAP can directly affect mitochondrial respiratory function and mtDNAcn. Treatment of cells with MDPs can counteract TRAP induced-effects. Lastly, we present evidence that suggests MDPs may be regulated in part by mitochondrial DNA methylation in humans.
Keywords: mitochondria, air pollution, particulate matter, DNA methylation, traffic, epigenetics
1. Introduction
Mitochondria are the cellular organelles that serve as central regulators of metabolism and oxidative stress. Dysfunctional mitochondria have been implicated in a variety of diseases including neurodegeneration and diabetes.(1–6) Mitochondrial DNA (mtDNA) was traditionally thought to only encode 13 protein coding genes, however, a novel class of bioactive, endogenous peptides that are encoded from small open reading frames within the mitochondrial genome has recently been discovered.(7–10) These mitochondrial derived peptides (MDPs) include HN, MOTS–c and SHLPs and have distinct biological activities on metabolism and cyto-protection as well as being altered in disease states (10–14), providing support for a network of active mitochondrial-encoded signals that act at the cellular and organismal level.
An emerging body of literature suggests that mitochondria may also contain machinery required to epigenetically modify mtDNA, such as mtDNA methylation, and affect its transcription.(15–17) This topic is controversial, as at least one study has suggested the secondary and tertiary structure of mtDNA can lead to artifacts in the measurement of mtDNA methylation, depending on the assay used.(16) Nevertheless, many nuclear-encoded mitochondrial genes are regulated by epigenetics such as DNA methylation.(18) Recent studies have also demonstrated the presence of methyltransferase including mtDNMT1, TET1 and TET2 activity in the mitochondria further suggesting the presence of mtDNA methylation.(19–21) Currently, the physiological role of mtDNA methylation is unknown; however the recent evidence for mtDNA methylation suggests we may hypothesize a role in disease risk, possibly by mediating risks conferred from environmental exposures.(22, 23)
Mitochondrial DNA is sensitive to damage by exogenous reactive oxygen sources. Mitochondrial oxidative damage, DNA copy number and DNA mutations have been studied in relation to both environmental exposures and disease outcomes. In particular, environmental pollutants such as traffic-related air pollution (TRAP), known to generate oxidative stress, have been associated with various forms of mitochondrial damage. Other air pollutants associated with the generation of oxidative stress include particulate matter (PM), carbon monoxide (CO), nitrogen dioxide (NO2), volatile organic compounds (VOCs), and polycyclic aromatic hydrocarbons (PAHs). Airborne particulate matter is defined by three size classes: coarse, PM10; <10μm diameter; fine, PM2.5; <2.5μm; and ultrafine or nano, PM0.2; <0.2μm. Both PM10 and PM2.5 can penetrate cells and damage the mitochondria, including disruption of structure and function and altered mtDNA copy number (mtDNAcn) in different tissues. (24–27) For example, in a study of mice, PM2.5 exposure induced insulin resistance and decreased mitochondrial count in visceral adipose tissue.(26) On the other hand, individual exposures to PM10 and coarse particles (PM10-PM1) showed higher mtDNAcn (24), which may be a compensatory mechanism against damaged mitochondria upon PM exposure. Benzo(a)pyrene (BaP), a polycyclic aromatic hydrocarbon (PAH) formed from incomplete combustion, is associated with mitochondrial release of cytochrome c, caspase-3 activation and neuronal apoptotic death as well as increased levels of reactive oxygen species (ROS) and lipid peroxidation within the mitochondria.(28–30) Studies are just beginning to evaluate associations between pollutants and potential changes in DNA methylation level within the mitochondria as a potential underlying mechanism. For instance, PM10 was associated with higher mtDNA methylation levels in peripheral blood (31) and PM2.5 was associated with higher mtDNA methylation levels in placental tissue but lower levels in peripheral blood, depending on the locus evaluated. (32, 33) PM2.5 also induced changes in methylation in mitochondrial-related nuclear genes and in mtDNA copy number. (34)
Mitochondria are one of the key players in the regulation of fetal programming and early development, as mitochondria are the primary energy producers of adenosine-5’-triphosphate (ATP) via oxidative phosphorylation. Given the important role for mitochondria in human disease, the increasing understanding of mito-regulatory mechanisms and the suggested associations between air pollutants and mitochondrial damage, we hypothesized that prenatal exposure to air pollution may be associated with mitochondrial dysfunction in the newborn and that MDPs might protect against these effects. We investigated the associations between mitochondrial respiration, mtDNA copy number, and MDP levels in response to air pollutant exposures using a combination of in vitro, in vivo and human studies. In in vitro and in vivo studies, we sought to demonstrate as proof of principle that pollutant exposures directly affect mitochondrial respiration, mtDNAcn and levels of MDPs in tissues previously implicated in pollutant-associated mitochondrial effects, such as neurons and liver. In a human population of pregnant women, we next sought to demonstrate similar associations between a wide variety of common air pollutant exposures on the potential for impaired mitochondrial function and MDPs in the newborn. We additionally sought to evaluate whether mtDNA methylation might correlate with MDP levels, thereby suggesting a role for mtDNA methylation in regulation of MDPs.
2. Materials and Methods
2.1. TRAP–UFP collection for in vitro and in vivo experiments
Ultrafine particulate matter (TRAP–UFP, <200 nm diameter) is a subfraction of PM2.5 collected from urban air in Los Angeles, California, near the CA–110 Freeway,(35) which represents a mix of fresh and aged ambient PM mostly from vehicular traffic.(36) Ultrafine PM is traditionally defined as particulates originating mostly from “fresh” emission sources and accounting for > 90% of the number-based particle concentrations. Recent studies in the Los Angeles Basin have shown that median mobility diameters in the inland valleys (downwind receptor areas) of the basin are in the 90–180 nm range in the summer months.(37) Therefore, for ultrafine PM measured in the Los Angeles Basin we define the particles as <.2 μm. The UFP was collected on Teflon filters and resuspended in ultrapure, deionized water by vortexing and sonication.(38) Water soluble metals and organic compounds are efficiently transferred from the filter collection medium into aqueous suspension used for exposures.(38) The chemical composition of TRAP–UFP used in these exposures has been reported previously. (39) The in vitro and in vivo doses are listed below.
2.2. In vitro assays
DNA from SH-SY5Y cells pretreated for 2 hours with 20μM MDPs (HNG, SHLP2, MOTS–c) followed by 24 hours treatment with the aqueous suspension of 10 ug/ml TRAP–UFP was extracted with a commercial kit (Qiagen, Valencia, CA, USA) and quantified by NanoDrop (Thermo Scientific, Wilmington, DE, USA). Mitochondrial copy number was estimated by real-time PCR (CFXConnect Real-Time System, Biorad, CA,USA) using two mtDNA targets (ND1, CYB) and two nuclear DNA targets (β–actin, 36B4) (IDT,CA, USA). The Q–PCR was performed in a 20 μL reaction mixture containing 10 μL SYBR Green, 50nM of each primer, and 20 ng of gDNA. gDNA was pretreated with HindIII to linearlize the circular mtDNA. The PCR reactions were subjected to a hot start at 98°C for 2 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing at 55°C for 30 seconds. The ratio of mtDNA to nuclear DNA was calculated by averaging the copy numbers of ND1/β–actin and CYB/36B4. The primers used for amplification of the human samples are listed in Table S1.
Cellular bioenergetics was determined by measuring oxygen consumption rate (OCR) of the cells with an XF–96 Flux Analyzer (Seahorse Biosciences, North Billerica, MA, USA). A seeding density of 20,000 cells per well was selected to allow both potential changes in OCR. The SH-SY5Y cells were pre-treated with 20μM MDPs (HNG (a potent analog of humanin S14G), SHLP2 and MOTS–c) for two hours followed by treatment with an aqueous suspension of 10 ug/ml TRAP–UFP or water for 24 hours. Assays were initiated by replacing the growth medium with 175 μL of XF assay medium (specially formulated, unbuffered Dulbecco’s modified Eagle’s medium for XF assays; Seahorse Bioscience) supplemented with 1mM sodium pyruvate and 25 mM glucose, pH 7.4. The cells were kept in a non–CO2-incubator for 60 minutes at 37°C before placement in the Analyzer. The basal OCR was measured using 3 cycles of 3 minute wait and 4 minute measure time. Each sample was measured in five to eight wells per condition and the results were averaged.
2.3. In vivo assays
Adult male C57BL/6 mice (N=12/group) were exposed to TRAP–UFP 5 hours per day, 3 days per week, for 10 weeks. Collected TRAP–UFP was reaerosolized, mixed with HEPA-filtered air, and delivered at a constant concentration (340 μg/m3). This dose was chosen as it represents a physiologically relevant human dose. The mice exposure duration of 150 hrs represents roughly 1% of their lifetime, and equivalently about 1 month of human exposure. During this exposure period, the PM concentration that would have resulted in the same PM dose per kg of body weight (2.5 mg/kg), is 33 μg/m3. This is a relevant PM exposure level, typical of PM2.5 concentrations in Los Angeles, and other urban areas of the US.(40) Control mice were exposed to only HEPA–filtered air. For the purpose of exposure, mice were transferred from home cages into sealed exposure chambers that allowed adequate ventilation and returned to home cages immediately after exposure. After 10 weeks of exposure, livers were collected to examine mitochondrial function, mtDNAcn, mtDNA oxidation and MDP levels.
Cellular bioenergetics of the chopped livers was determined by measuring oxygen consumption rate (OCR) of the cells with an XF–96 Flux Analyzer (Seahorse Biosciences, North Billerica, MA, USA). Assays were initiated by replacing the growth medium with 175 μL of XF assay medium (specially formulated, unbuffered Dulbecco’s modified Eagle’s medium for XF assays; Seahorse Bioscience) supplemented with 1mM sodium pyruvate and 25 mM glucose, pH 7.4. The tissues were kept in a non–CO2-incubator for 60 minutes at 37°C before placement in the Analyzer. The basal OCR was measured using 3 cycles of 3 minute wait and 4 minute measure time. Each sample was measured in five to eight wells per condition and the results were averaged.
2.4. Human Studies
We evaluated the associations between air pollutants and mitochondrial MDPs, mtDNA methylation and copy number in a human population that included 82 mother-infant pairs from the University of Southern California Maternal and Children Health (MACHS) birth cohort study. The 82 pairs are a subset of the larger MACHS study of 232 mother-child pairs who were recruited on the labor and delivery ward at the Los Angeles County + University of Southern California (LAC+USC) Medical Center from September 2012 to August 2015. Written informed consent was obtained from each pregnant woman prior to any testing. Exclusion criteria included <18 years of age, HIV positive status, physical, mental, or cognitive disabilities that prevented participation, current incarceration, or multiple pregnancy. A history of socio-demographic variables was obtained at study entry. Medical record information pertaining to pregnancy complications and delivery was also obtained. Maternal information included maternal age, gestational age at delivery, race, parity, stress level during pregnancy, family income, maternal education and maternal body mass index (BMI) prior to pregnancy, which was calculated based on mother’s pre-pregnancy weight and height. Infant birth weight was extracted from hospital medical records. Cord blood was collected at the time of delivery. The assessment of MDPs was only performed in 82 subjects chosen at random as part of a pilot project. Therefore all statistical analyses were restricted to this subset to ensure continuity across all statistical associations.
Traffic related air pollutant (TRAP) and ambient air pollutant (AAP) exposures including PM10, PM2.5, O3 and NO2, were estimated based on participant’s residential addresses reported at study entry. Exposure to local TRAP was estimated using modeled nitric oxides (NOx) at homes by applying the CALINE4 (“California Line Source”) dispersion model.(41) The CALINE4 dispersion model is a dispersion model well suited for estimating vehicle emissions concentrations downwind of roadways. It uses meteorological data, roadway geometry, traffic volumes, and vehicle emission factors. We have utilized CALINE4 in many studies to characterize spatial patterns of exposure to traffic-related pollutants.(42) TRAP and AAP concentrations for the 9 months prior to delivery were calculated and examined at prenatal exposures. For AAP, street-level residential addresses of participants were standardized and geocoded at the parcel level and match codes were obtained using the Texas A&M Geocoder (http://geoservices.tamu.edu/Services/Geocode/). Addresses that did not match to a parcel centroid were corrected based on the best available knowledge of the participant’s residence location. Using routine air monitoring data collected daily in California between 2011 and July 2015, and available from the US EPA’s Air Quality System, we estimated the daily ambient concentrations at the participants’ geocoded residence location using inverse-distance-squared weighted interpolation. Ambient exposures estimated at each residence were obtained using spatial interpolation from air quality monitoring stations nearest to the participant’s residence at the finest geographic resolution possible (usually parcel-level) using inverse distance-squared weighting.(43)
Benzo[a]pyrene (BaP) is bio–activated to BaP diol–epoxides (BPDEs) that can bind to hemoglobin to form protein adducts. Disruption of the tertiary structures of the protein releases BaP tetrol metabolites, which are used as a quantitative measurement of BaP–Hb adducts.(44–46) RBCs were isolated from 2 ml of blood, then washed three times with PBS and lysed with 10−4 M EDTA (pH7.5). After centrifugation, the supernatant was transferred to a 50 ml centrifuge tube and ice cold acetone was used to precipitate hemoglobin. The precipitate was washed three times with acetone and air dried, followed by hydrolysis with 1 N sodium hydroxide to release BaP tetrol. The hydrolyzed sample was then extracted with ethyl acetate, and analyzed by LC–APCI–MS/MS. The detection limit for Bap-Hb adduct was 0.312 pmol/g Hb.
A 15–cc cord blood sample for each participant was collected by hospital providers in 1 EDTA tube for plasma and DNA isolation. The blood sample was stored at room temperature until transportation within 24 hours to the molecular biology laboratory at the Southern California Environmental Health Sciences Center, where the sample was then processed. The EDTA tube was centrifuged at 1500 xg for 10 minutes. The buffy coat was collected and lysed for DNA extraction. Circulating levels of MDPs including humanin (HN), small humanin-like peptides (SHLPs) and mitochondrial open-reading-frame of the 12S rRNA–c (MOTS–c) were measured by an in-house sandwich ELISAs seperately. For the HN assay, plasma was extracted with 90% acetonitrile and 10% 1N HCl.(7) Briefly, 200μl of extraction reagent was added to 100 μl of plasma gently mixed and incubated at room temperature. The supernatant was removed and dried by SpeedVac after centrifuge. The dried extracts were reconstituted with PBS and then used for ELISA. For the HN measurement, synthetic HN was used as a standard within a range of 0.1 ng/ml to 50ng/ml. 96-well microtiter plates were coated with a HN capture antibody in 50 mM sodium bicarbonate buffer on a shaker. The plates were washed and blocked with Superblock buffer. Standards, controls or extracted samples and pre-titered detection antibody were added to the appropriate wells and incubated overnight. The absorbance was read at 490 nm on a plate spectrophotometer following streptavidin–HRP and OPD incubation. The SHLPs and MOTS–c were measured using a similar procedure with capture and detection antibodies specifically against SHLPs (8) or MOTS–c peptides. (10)
To measure DNA methylation, DNA was extracted from the buffy coat using the QIAamp DNA Blood Midi Kit (Qiagen) and then bisulfite-treated using the EZ–96 DNA Methylation Kit (Zymo Research) according to the manufacturer’s instructions. Mitochondrial DNA methylation was assayed using Pyrosequencing. Methylation analyses were performed by bisulfite-PCR. Pyrosequencing assays were performed using the HotMaster Mix (Eppendorf, Hamburg, Germany) and the Pyrosequencing (PSQ) HS 96 Pyrosequencing System (Qiagen, Inc) as described in previous work (Byun et al. (47)). The Pyrosequencing instrument includes built-in controls for assessing completion of bisulfite conversion. The assays measure methylation in three mtDNA regions: the mitochondrial encoded transfer RNA Phenylalanine (MT–TF), the 12S ribosomal RNA (MT–RNR1) and the D-loop control region. The polymerase chain reaction (PCR) and pyrosequencing primers from Byun et al.(48) were used for measuring MT–TF methylation only. For the MT–RNR1 region, the new set of PCR primers was modified with biotin-labeled forward primer, although both forward and reverse primer sequences were same as the primers for MT–TF. One CpG in each of MT–TF and MT–RNR1, 3 CpGs from the D–loop region, and one CpG from the light chain in D–loop region (LDLR2) (49) were analyzed. Nine samples assayed or D–loop did not pass the Pyrosequencer QC and were removed from the analysis. Primers are shown in Table S2. CpG loci evaluated in the mitochondrial genome are shown in Figure S1.
Relative mitochondrial copy number (mtDNAcn) was measured by qPCR assay by determining the ratio of mitochondrial (Mt) copy number to a single copy gene (human [beta] globin: hbg) number in experimental samples relative to a reference as described in the in vitro experiments. All human samples were run in triplicate. Standard deviations for the threshold cycle (Ct) less than or equal to 0.25 were acceptable. A control DNA sample was included in each qPCR. The reference DNA, which is pooled DNA from all MACHs samples, was run in duplicate. A fresh standard curve, which ranged from 20 ng/μl to 0.625 ng/μl was generated in each Mt and hbg qPCR run. The R2 for each standard curve was repeated if it was less than 0.99.
3. Statistical Methods
For in vitro assays, differences between pollution treated and control cells were calculated using oneway ANOVA. For in vivo experiments, differences between treated and control mice were compared using a t-test. In the human studies, we first calculated Spearman’s correlation coefficients to evaluate relationships between air pollutant exposures, MDP levels, mtDNA copy number, and mtDNA methylation in cord blood. Air pollutant exposures were dichotomized into high and low values to test mean differences and were scaled to a 2-standard deviation range in linear regression models.
In the human study we first evaluated the normality of distributions of methylation, mtDNA copy number and MDPs using the Shapiro-Wilk test for normality test. To estimate the association between prenatal air pollutant exposures and MDPs, we fitted linear regression models for each pollutant and adjusted for maternal age, gestational age, ethnicity, maternal smoking and season of birth (defined as warm season if baby was born between March and September, or cool season otherwise). These covariates were chosen for inclusion in models based on a priori knowledge for their potential to act as confounders in studies of air pollution. Parity, date of delivery, maternal education and income level were also evaluated as potential confounders in these models but were subsequently removed because they did not change the observed results or conclusions and did not meet the definition of a confounder in this analysis. To test the association between prenatal air pollutants and mtDNA copy number, linear mixed effects models were fitted in which mtDNA assay plate number was included as random effect. We applied similar linear regression models to test the association between mtDNA methylation and MDPs, adjusting for maternal age, gestational age, ethnicity, maternal smoking and gestational diabetes mellitus (evaluated based on results from OGGT and GCT tests in the medical record). For those outcomes found to have non-normal distributions, we conducted sensitivity tests in which we log-transformed the outcomes and compared the results from linear regression models to models with untransformed variables.
Statistical analyses were performed using SAS (Statistical Analysis System) version 9.4 (SAS Institute, Cary, NC) and R version 3.3.1 software.
4. Results
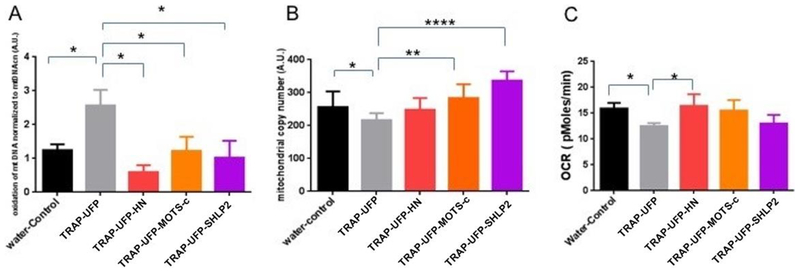
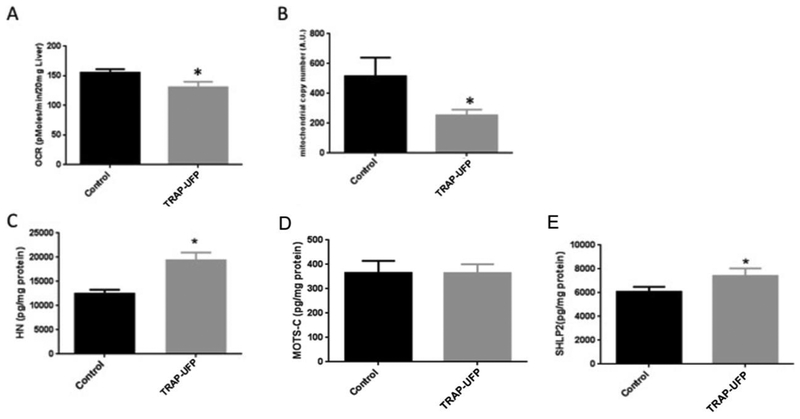
We first evaluated whether exposure to TRAP–UFP affected mitochondrial function in vitro and in vivo. 24 hour TRAP–UFP exposure increased oxidation of mtDNA, decreased mitochondrial consumption rate, and decreased mtDNA copy number (mtDNAcn) in SH–SY5Y cells (Figure 1). Addition of MDPs HN, MOTS–c and SHLP2 rescued these effects to varying degrees. Addition of all three MDPs rescued the effects on oxidation of mtDNA, whereas addition of only HN raised the OCR to baseline levels. Also, cells exposed to TRAP–UFP decreased the mtDNAcn which was reversed by addition of MOTS–c and SHLP2 but not HN. In the mouse model, we investigated whether TRAP–UFP altered OCR, MDP levels and mtDNAcn. Male mice exposed to TRAP–UFP for 150 hours over 10 weeks had lower OCR, lower mtDNAcn and higher HN and SHLP2 levels in mouse liver (Figure 2). Together, these data suggest MDPs may protect cells against oxidative damage from air pollution, and the elevated levels of MDPs in mouse liver could be a compensatory mechanism against air pollution. We next investigated the effects of a variety of air pollutant exposures in a human population (MACHS study) on MDPs and mtDNAcn in circulating blood cells. We also evaluated mtDNA methylation in locations of the genome potentially relevant to the regulation of MDPs. Baseline characteristics of the 82 mother-infant pairs and distribution of traffic-related air pollution (TRAP) and ambient air pollutant (AAP) exposures are shown in Tables 1 and S3, for the subset of 82 and for the entire cohort. Mothers were predominantly Hispanic (91%) and low-income, with 64% of them making less than $30,000 a year. Infant sex was evenly distributed and nearly half of the mothers were nulliparous. Ten percent of women had gestational diabetes and only 5% had ever smoked at any time in their pregnancy. The subset of 82 mothers included in this study was reflective of the MACHs population as a whole. Mean levels of AAP pollutants and BaP in the substudy were representative of the parent cohort with the exception of O3. Prenatal TRAP tended to be lower in the substudy population. Prenatal pollutants were moderately correlated with one another (Table S4). Prenatal NO2 was positively correlated with PM2.5 (r=0.4) and negatively correlated with O3 (r= −0.7). PM2.5 and O3 were negatively correlated (r= −0.6), reflecting patterns typically observed in ambient air pollutants in southern California.
Figure 1: In vitro PM2.5 nanoparticle exposure impairs mitochondrial function which is ameliorated by mitochondrial peptides.
SH–SY5Y cells were exposed to 10-μg/ml solubilized nPM -nanoparticles for 24 hours alone, or in the presence of 20M MDPs (HN, μ2.5MOTS–c and SHLP2). (A) nPM nanoparticles caused an increase in oxidation of mtDNA (p<0.01). This effect was rescued by all three 2.5MDPs. (B) Mitochondrial copy number measured by qPCR decreased with nPM nanoparticles exposure (p<0.01). This effect was 2.5rescued by MOTS–c and SHLP2. (C) nPMexposure decreased mitochondrial consumption rate measured by Seahorse XF96 Analyzer 2.5 (p<0.01). This effect was rescued by HN.
Figure 2: In vivo TRAP–UFP exposure impairs mitochondrial function and increases MDP levels in murine liver.
C57Bl/6 male mice were exposed to TRAP–UFP or Control air for 10 weeks and liver was collected to examine the mitochondrial function, mtDNAcn, mtDNA oxidation and MDP levels. (A) TRAP–UFP exposure decreased mitochondrial oxygen consumption rate in ex vivo hepatic explants measured by Seahorse XF96 Analyzer (p<0.01). (B) Mitochondrial copy number in liver measured by qPCR decreased with TRAP–UFP exposure after 10 week exposure (p<0.01). TRAP–UFP increased HN (C), did not affect MOTS–C (D) and increased SHLP2 (E) levels in liver (p<0.01).
Table 1.
Descriptive characteristics of the Maternal and Children Health study and the subset of 82 mother-infant pairs
| Sub-study of MDPs (N=82) | MACHS parent cohort (N=232) | ||||
|---|---|---|---|---|---|
| N | % | N | % | p-value | |
| Hispanic ethnicity | 75 | 91.5 | 203 | 87.5 | 0.33 |
| Male infant | 42 | 51.2 | 115 | 50.4 | 0.90 |
| Maternal smoking during pregnancy | 4 | 4.9 | 21 | 9.1 | 0.23 |
| Family Income | |||||
| Less than $15,000 | 24 | 29.3 | 66 | 28.5 | 0.38 |
| $15,000 to $29,999 | 29 | 35.4 | 64 | 27.6 | |
| $30,000 to $49,999 | 10 | 12.2 | 22 | 9.5 | |
| $50,000 or more | 3 | 3.6 | 7 | 3.0 | |
| Caesarean Section | 30 | 37.8 | 82 | 36.4 | 0.59 |
| Parity | |||||
| 0 | 39 | 47.6 | 115 | 50.4 | 0.85 |
| 1 | 23 | 28.1 | 57 | 25.0 | |
| 2+ | 20 | 24.4 | 27 | 11.8 | |
| Pre-Pregnancy Weight | |||||
| Underweight | 26 | 31.7 | 59 | 25.4 | 0.59 |
| Normal | 16 | 19.5 | 54 | 23.3 | |
| Overweight | 22 | 26.8 | 57 | 24.6 | |
| Obese | 18 | 22.0 | 59 | 25.4 | |
| Gestational Diabetes (GDM) | |||||
| Normal | 44 | 53.0 | 137 | 59.1 | 0.97 |
| Intolerance | 2 | 2.4 | 5 | 2.2 | |
| GDM | 5 | 6.0 | 15 | 6.5 | |
| Mean | SD | Mean | SD | p-value | |
| Gestational age (weeks) | 38.7 | 1.6 | 38.5 | 2.1 | 0.42 |
| Maternal age | 27.5 | 7.1 | 27.6 | 6.5 | 0.89 |
| Birthweight (grams) | 3222.3 | 476.4 | 3193.0 | 544.3 | 0.67 |
P-values for categorical characteristics derived using Pearson chi-square test unless otherwise noted; p-values for continuous characteristics derived using t-test. Missing values were not included.
The distribution of MDPs, mitochondrial copy number and mitochondrial DNA methylation is shown in Table 2. DNA methylation levels in the mitochondrial genome were largely low. LDLR2 showed the highest levels of methylation in whole blood, with a mean of 10.7% (SD 4%). mtDNA methylation levels were not highly correlated with one another, with Spearman correlation coefficients ranging from 0.24 to 0.29 (Table S5). MDP levels show a large inter-individual variation, with HN having the greatest dynamic range in values in cord blood. Several MDPs in whole blood were minimally correlated with one another (Table S6). HN was correlated with SHLP2 (r = 0.46) and SHLP2 with MOTS–c (r=0.26), respectively. mtDNAcn was not highly correlated with MDPs or with methylation levels.
Table 2.
Descriptive statistics of MDPs, mtDNAcn and mtDNA methylation
| N | Minimum | Q1 | Median | Mean | SD | Q3 | Maximum | |
|---|---|---|---|---|---|---|---|---|
| mtDNA copy number (unitless) | 82 | 0.2 | 0.4 | 0.6 | 0.7 | 0.4 | 0.8 | 2.00 |
| Peptides(pg/ml) | ||||||||
| HN | 73 | 243 | 622 | 1066 | 1134 | 711 | 1500 | 4813 |
| MOTS-c | 80 | 90 | 325 | 428 | 437 | 160 | 557 | 834 |
| SHLP2 | 82 | 248 | 491 | 527 | 611 | 257 | 636 | 1658 |
| Whole blood methlyation % | ||||||||
| MT-TF | 82 | 0 | 0.5 | 0.9 | 0.8 | 0.5 | 1.1 | 2.1 |
| MT-RNR1 | 82 | 1.1 | 2.2 | 2.5 | 2.6 | 0.7 | 2.8 | 5.7 |
| average of LDLR2 | 82 | 5.1 | 8.5 | 10.2 | 10.7 | 4 | 12.1 | 33.8 |
| average of D-loop | 75 | 0.7 | 2.2 | 2.6 | 2.9 | 1.4 | 3.3 | 11.3 |
4.1. Association between air pollution and mtDNAcn and MDPs
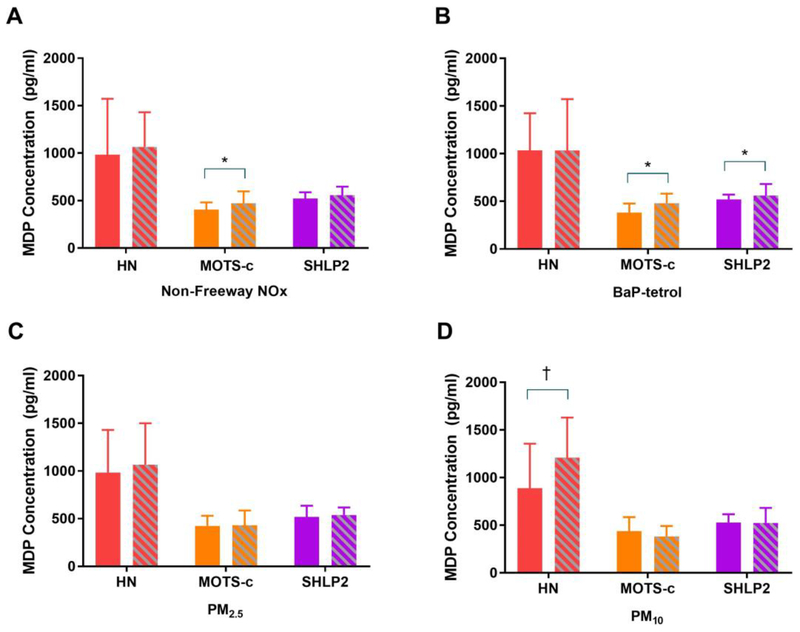
Subjects exposed to high non-freeway TRAP levels had significantly higher MOTS–c levels, subjects with high PM10 exposure had higher HN, and subjects with high BaP–tetrol, a derivative of Benzo(a)pyrene measured in the cord blood, had higher SHLP2 and marginally higher MOTS–c levels compared to the unexposed group (Figure 3). PM2.5 showed a similar but non-significant difference. BaP–tetrol was also associated with higher mtDNA copy number (β= 0.14 per 2 SD change BaP–tetrol, p=0.03) however other pollutants were not associated with mtDNA copy number. In linear models, subjects with higher non-freeway TRAP had higher levels of all MDPs except HN (increase ranged from 88.6 to 124.7pg/ml per 2SD pollutant). A 2 SD higher BaP-tetrol was also associated with 167.6 pg/ml higher levels of SHLP2 (Figure S2).
Figure 3. Associations between traffic-related air pollutants, ambient air pollutants, BaP tetrol and MDPs.
(A) Non-Freeway NOx (B) BaP tetrol (C) PM2.5 (D) PM10. Median (IQR) of MDPs are shown by high-exposed and low-exposed group. Solid color represents low-exposed group whereas color in shade represents high-exposed group. Cutoffs for high-exposed group and low-exposed group are 2.23 ppb for non-freeway NOx, 0.50 pmol/g for BaP tetrol, 12.76 ug/m3 for PM2.5, and 30.39 ug/m3 for PM10. *p-value <0.05, † p-value<0.10.
4.2. Associations between peptides and mtDNA methylation
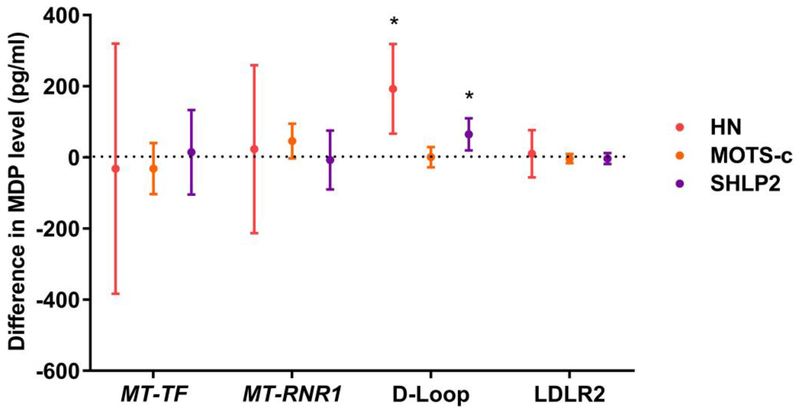
Higher DNA methylation of the D–Loop in cord blood was associated with higher HN and SHLP2 levels whereas higher methylation of MT–RNR1 was associated with higher MOTS–c levels (which is encoded from the 12S-rRNA/MT–RNR1 region) (Figure 4), after adjustment for maternal age, gestational age, ethnicity, maternal smoking, and gestational diabetes. For example, a 1% higher methylation level of the D-Loop was associated with a 193 pg/ml higher HN level (p=0.003) and a 65 pg/ml higher SHLP2 level (p=0.01). A 1% higher MT–RNR1 methylation level at CpG 7 was marginally associated with a 46 pg/ml higher MOTS–c level (p=0.07) (Table S7).
Figure 4. Association between mtDNA methylation and MDP levels in whole blood.
Estimated difference in MDP level is shown for a 1% change in methylation with 95% confidence interval. Models adjusted for maternal age, gestational age, ethnicity, maternal smoking during pregnancy, and GDM. *p-value <0.05.
4.3. Sensitivity analyses
We conducted sensitivity analyses in which we modeled MDPs and mtDNA methylation values using log-transformed variables given that most of them were significantly right-skewed (Tables S8 and S9);however, our conclusions were unchanged. We conducted sensitivity analyses in which our main models for associations between MDPs with mtDNA methylation and air pollutants with MDPs were additionally adjusted for date of delivery, parity, income and maternal education (Tables S10 and S11). Adjustment for these additional variables did not materially influence our results.
5. Discussion
Our in vitro and in vivo experimental and human data indicates that air pollutants can directly affect mitochondrial respiratory function and mtDNA copy number. Moreover, treatment of cells with HN or MOTS–c can counteract air pollutant induced-effects. We also show that both MDP and mtDNAcn levels in mouse and newborn samples are affected by pollutant exposures. Lastly, we present evidence that suggests MDPs may be regulated in part by mitochondrial DNA methylation in humans. DNA methylation of two distinct regions of the D-Loop in the mitochondria genome was associated with levels of several MDPs. DNA methylation in MTRNR1 is associated with MOTS–c level, an MDP that is encoded from the 12S-rRNA/MTRNR1 region.
Mitochondria are sensitive to exogenous stress from prenatal air pollutant exposures but different exposures have elicited different responses, including decreases in energy metabolism resulting in reduced sperm motility(50), altered mtDNAcn in offspring (51, 52), fetal growth restriction and placental mitochondrial impairment (53). In our in vitro and and in vivo work, TRAP–UFP exposure increased mtDNA oxidation and decreased mitochondrial consumption rate. TRAP–UFP exposure also decreased mtDNAcn whereas in our human population we observed no associations with PM. One explanation for these differences may be in the chemical composition of the particles themselves. TRAP–UFP has a higher level of redox active species such as organic carbon and transition metals compared to PM2.5 and PM10 which leads to differences in oxidation potential(54). Our results are similar to results found by others in short term exposure studies and in studies of pregnancy (31–33, 55). The prevailing theory is that compensatory mitochondria biogenesis can buffer an intracellular reactive oxygen species (ROS) challenge as part of an adaptive stress response (56, 57). However, persistent oxidative stress may eventually overwhelm the adaptive response system and lead to mitochondrial DNA depletion (58). Previous studies have observed similar responses, whereby both an initial upregulation of energy producing pathways may occur but also an inhibition of mitochondrial biogenesis upon exposure to repeated low-level particulate matter (59, 60). Given this fine balancing act, dose of a given pollutant, type of pollutant and maintenance of a steady state may be particularly important. Variations to any of these in a given study may lead to conflicting sets of results. For instance, we observed a positive association with BaP–tetrol and mtDNAcn, which is seemingly contradictory to the proposed hypothesis. However the exposures in vitro were acute whereas the exposures in the human study were chronic, and BaP tetrol has not previously been investigated with respect to mtDNAcn and may in part explain differences in results. Alternatively, our sample size in the population study was small and we may have had limited power to detect small differences.
To our knowledge, associations between air pollutants and effects on MDPs have not been evaluated in humans. In this study, we found that prenatal TRAP and BaP-tetrol were associated with higher SHLP2 and MOTS–c. The in vivo and in vitro experiments also demonstrated higher MDP levels in response to pollutant exposure as well as the ability of MDPs to rescue the negative effects of PM on mitochondrial respiration. These results suggest that MDPs are broadly up-regulated to protect against pollutant-induced mitochondrial damage. In fact, the upregulation of MDPs in response to a cellular insult has been suggested before for muscle cells responding to an energy crisis.(61) Other studies have shown that humanin protects against various pathological conditions including chemotherapy, neuronal cell death, and oxidative stress. Along with the humanin’s cytoprotective roles, the up-regulation of humanin is a compensatory mechanism against pollutant-induced mitochondrial damage.
Lastly, we found that higher DNA methylation of the D–Loop in cord blood was associated with higher HN and SHLP2 levels. Surprisingly, the exact purpose of the D–Loop in mitochondrial function remains unknown, although several theories exist, including that it is an intermediate form of mitochondria replication, that it serves to bring together multiple copies of mtDNA, and that it is involved in dNTP metabolism (62). The two regions we evaluated were 382 nucleotides apart, residing in different putative functional regions of the D–Loop, with one in the hypervariable 2 region and the other in the MTTF binding site. Our data suggest that D-Loop region of the mitochondria may have a role to play in regulating transcription of MDPs. It may be that increased methylation affects overall transcription of the mtDNA, resulting in higher levels of MDPs. However, we also observed that higher methylation of a CpG in MTRNR1 was associated with higher MOTS–c levels. Given that MOTS–c is transcribed from an open reading frame within MTRNR1, DNA methylation at the CpG locus may affect MOTS–c transcription. This result also suggests that there is a non-canonical mitochondrial transcription mechanism in addition to the canonical polycistronic transcription model.
We acknowledge several limitations in the present study. In vitro and in vivo experiments do not reflect the totality of biological systems in humans and therefore results cannot always be extrapolated. Air pollutant exposures used in the in vivo and in vivo experiments differed from those measured in the population-based study, though all were derived from real traffic-related pollution in the Los Angeles air basin. Nonetheless, the fact that we observed consistent directions of association on MDP levels despite differences in exposure is a strength of the study and suggests a common mechanism that is not sensitive to particulate matter particle size. In our populatin study, PM2.5 and PM10 were negatively correlated which is atypical. This negative correlation was driven solely by correlation within the year 2015. The year 2015 was a strong El Niño year in California. Studies have previously reported that strong El Niño conditions may result in changes in PM2.5 chemical composition and disrupted correlation patterns.(63–65) Therefore, we further adjusted calendar year as a binary variable (2015 vs 2014) in ambient air pollution models. Only three regions of mtDNA were selected and assayed in the present study. Our findings may not extrapolate to the entire mtDNA genome. Another challenge that may affect mtDNA methylation detection is presence of nuclear-mitochondrial sequences (Numts), or pseudogenes in the nuclear genome that do not transcribe. Current bisulfite treatment of total genomic DNA for methylation analysis may not be able to distinguish mtDNA sequences and Numts although we designed primers specific to the mtDNA sequence and checked them against the nuclear genome for potential overlap which greatly reduces this likelihood.
5.1. Conclusion
The combination of in vitro, in vivo and human studies suggest that ultrafine particles and traffic-related air pollution can affect mitochondrial respiratory function, mitochondrial copy number, and MDP levels. Specifically, treatment of cells with MDPs can counteract air pollutant induced-effects and in mice and humans MDP levels are elevated, suggesting a protective role for MDPs against pollutant-inducing mitochondrial damage. Lastly, MDP levels were correlated with mitochondrial DNA methylation in newborns, suggesting a potential regulatory mechanism which warrants further investigation.
Supplementary Material
Grant funding
We would like to acknowledge the following funding sources for this work: to CB: P30ES007048, R21ES025870; to PC: R01AG061834, P01AG034906, DODPC160353, U54CA233465, and an AFAR BIG AWARD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
Dr. Cohen is a stockholder and SAB member of CohBar Inc.
References
- 1.Berdanier CD, Linking mitochondrial function to diabetes mellitus: an animal’s tale. Am J Physiol Cell Physiol 293, C830–836 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Rabol R, Boushel R, Dela F, Mitochondrial oxidative function and type 2 diabetes. Appl Physiol Nutr Metab 31, 675–683 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Schrauwen-Hinderling VB, Kooi ME, Schrauwen P, Mitochondrial Function and Diabetes: Consequences for Skeletal and Cardiac Muscle Metabolism. Antioxid Redox Signal 24, 39–51 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Chowdhury SK, Dobrowsky RT, Fernyhough P, Nutrient excess and altered mitochondrial proteome and function contribute to neurodegeneration in diabetes. Mitochondrion 11, 845–854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenaz G, Baracca A, Fato R, Genova ML, Solaini G, Mitochondrial Complex I: structure, function, and implications in neurodegeneration. Ital J Biochem 55, 232–253 (2006). [PubMed] [Google Scholar]
- 6.McInnes J, Insights on altered mitochondrial function and dynamics in the pathogenesis of neurodegeneration. Transl Neurodegener 2, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin YP et al. , Pharmacokinetics and tissue distribution of humanin and its analogues in male rodents. Endocrinology 154, 3739–3744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobb LJ et al. , Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging (Albany NY) 8, 796–809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C, Kim KH, Cohen P, MOTS–c: A novel mitochondrial-derived peptide regulating muscle and fat metabolism. Free radical biology & medicine 100, 182–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C et al. , The mitochondrial-derived peptide MOTS–c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 21, 443–454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahboobi H, Golmirzaei J, Gan SH, Jalalian M, Kamal MA, Humanin: a possible linkage between Alzheimer’s disease and type 2 diabetes. CNS Neurol Disord Drug Targets 13, 543–552 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Matsuoka M, Protective effects of Humanin and calmodulin-like skin protein in Alzheimer’s disease and broad range of abnormalities. Mol Neurobiol 51, 1232–1239 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Xiao J, Kim SJ, Cohen P, Yen K, Humanin: Functional Interfaces with IGF-I. Growth Horm IGF Res 29, 21–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen K, Lee C, Mehta H, Cohen P, The emerging role of the mitochondrial-derived peptide humanin in stress resistance. J Mol Endocrinol 50, R11–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Aquila P, Montesanto A, Guarasci F, Passarino G, Bellizzi D, Mitochondrial genome and epigenome: two sides of the same coin. Frontiers in bioscience (Landmark edition) 22, 888–908 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Mechta M, Ingerslev LR, Barres R, Methodology for Accurate Detection of Mitochondrial DNA Methylation. J Vis Exp, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stimpfel M, Jancar N, Virant-Klun I, New Challenge: Mitochondrial Epigenetics? Stem Cell Rev 14, 13–26 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Matilainen O, Quiros PM, Auwerx J, Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress. Trends Cell Biol, (2017). [DOI] [PubMed] [Google Scholar]
- 19.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM, DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A 108, 3630–3635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzitoyeva S, Chen H, Manev H, Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol Aging 33, 2881–2891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao D, Zhu B, Sun H, Wang X, Mitochondrial DNA Methylation and Related Disease. Adv Exp Med Biol 1038, 117–132 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Feng S, Xiong L, Ji Z, Cheng W, Yang H, Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol Med Rep 6, 125–130 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Chinnery PF, Elliott HR, Hudson G, Samuels DC, Relton CL, Epigenetics, epidemiology and mitochondrial DNA diseases. Int J Epidemiol 41, 177–187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gualtieri M et al. , Airborne urban particles (Milan winter-PM2.5) cause mitotic arrest and cell death: Effects on DNA, mitochondria, AhR binding and spindle organization. Mutat Res 713, 18–31 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Delgado-Buenrostro NL et al. , Decrease in respiratory function and electron transport chain induced by airborne particulate matter (PM10) exposure in lung mitochondria. Toxicol Pathol 41, 628–638 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Xu X et al. , Long-term exposure to ambient fine particulate pollution induces insulin resistance and mitochondrial alteration in adipose tissue. Toxicol Sci 124, 88–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou L et al. , Airborne particulate matter and mitochondrial damage: a cross-sectional study. Environ Health 9, 48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie JS, Zhang HM, Zhao J, Liu HJ, Niu Q, Involvement of mitochondrial pathway in benzo[a]pyrene-induced neuron apoptosis. Hum Exp Toxicol 33, 240–250 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Sobinoff AP, Pye V, Nixon B, Roman SD, McLaughlin EA, Jumping the gun: smoking constituent BaP causes premature primordial follicle activation and impairs oocyte fusibility through oxidative stress. Toxicol Appl Pharmacol 260, 70–80 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Ko CB et al. , Benzo(a)pyrene-induced apoptotic death of mouse hepatoma Hepa1c1c7 cells via activation of intrinsic caspase cascade and mitochondrial dysfunction. Toxicology 199, 35–46 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Byun HM et al. , Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol 10, 18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen BG et al. , Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: An ENVIRONAGE birth cohort study. Epigenetics 10, 536–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byun HM et al. , Effects of Air Pollution and Blood Mitochondrial DNA Methylation on Markers of Heart Rate Variability. J Am Heart Assoc 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong J et al. , B vitamins attenuate the epigenetic effects of ambient fine particles in a pilot human intervention trial. Proc Natl Acad Sci U S A, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra C, Kim S, Shen S, Sioutas C, Design and evaluation of a high-flow rate, very low pressure drop impactor for separation and collection of fine from ultrafine particles. Journal of Aerosol Science 33, 735–752 (2002). [Google Scholar]
- 36.Ning Z et al. , Daily variation in chemical characteristics of urban ultrafine aerosols and inference of their sources. Environ Sci Technol 41, 6000–6006 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Sioutas C, Delfino RJ, Singh M, Exposure assessment for atmospheric ultrafine particles (UFPs) and implications in epidemiologic research. Environ Health Perspect 113, 947–955 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan TE et al. , Glutamatergic neurons in rodent models respond to nanoscale particulate urban air pollutants in vivo and in vitro. Environ Health Perspect 119, 1003–1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodward NC et al. , Traffic-related air pollution impact on mouse brain accelerates myelin and neuritic aging changes with specificity for CA1 neurons. Neurobiol Aging 53, 48–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. S et al. , Spatial and Temporal Variability of Sources of Ambient Fine Particular Matter (PM2.5) in California. Atmospheric Chemistry and Physics 14, 12085–12097 (2014). [Google Scholar]
- 41.Benson PE, A Review of the Development and Application of the Caline3 and Caline4 Models. Atmos Environ BUrb 26, 379–390 (1992). [Google Scholar]
- 42.Franklin M et al. , Predictors of intra-community variation in air quality. J Expo Sci Environ Epidemiol 22, 135–147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong DW, Yuan L, Perlin SA, Comparison of spatial interpolation methods for the estimation of air quality data. J Expo Anal Environ Epidemiol 14, 404–415 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Padros J, Pelletier E, In vivo formation of (+)-anti-benzo[a]pyrene diol-epoxide-plasma albumin adducts in fish. Mar Environ Res 50, 347–351 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Frank S, Renner T, Ruppert T, Scherer G, Determination of albumin adducts of (+)-anti-benzo[a]pyrene-diolepoxide using an high-performance liquid chromatographic column switching technique for sample preparation and gas chromatography-mass spectrometry for the final detection. J Chromatogr B Biomed Sci Appl 713, 331–337 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Ragin AD, Crawford KE, Etheredge AA, Grainger J, Patterson DG Jr., A gas chromatography-isotope dilution high-resolution mass spectrometry method for quantification of isomeric benzo[a]pyrene diol epoxide hemoglobin adducts in humans. J Anal Toxicol 32, 728–736 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Byun HM et al. , Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 18, 4808–4817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.P. T Byun H, Motta V, Hou L, Nordio F, Apostoli P, Bertazzi P, Baccarelli A, Effects of airborne polutants on mitochondrial DNA methylation. Particle and Fibre Toxiocology 10, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.G. B Armstrong DA, Blair BA, Guerin DJ, Litzky JF, Chavan NR, Pearson KJ, Marsit CJ, Maternal Smoking During Pregnancy is Associated with Mitochondrial DNA Methylation. Environmental Epigenetics 2, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J et al. , PM2.5 induces male reproductive toxicity via mitochondrial dysfunction, DNA damage and RIPK1 mediated apoptotic signaling pathway. Sci Total Environ 634, 1435–1444 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Brunst KJ et al. , Prenatal particulate matter exposure and mitochondrial dysfunction at the maternal-fetal interface: Effect modification by maternal lifetime trauma and child sex. Environ Int 112, 49–58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clemente DB et al. , Prenatal Ambient Air Pollution, Placental Mitochondrial DNA Content, and Birth Weight in the INMA (Spain) and ENVIRONAGE (Belgium) Birth Cohorts. Environ Health Perspect 124, 659–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fetterman JL, Sammy MJ, Ballinger SW, Mitochondrial toxicity of tobacco smoke and air pollution. Toxicology 391, 18–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saffari A, Daher N, Shafer MM, Schauer JJ, Sioutas C, Global perspective on the oxidative potential of airborne particulate matter: a synthesis of research findings. Environ Sci Technol 48, 7576–7583 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Janssen BG et al. , Placental mitochondrial DNA content and particulate air pollution during in utero life. Environ Health Perspect 120, 1346–1352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong J et al. , Traffic-Related Air Pollution, Blood Pressure, and Adaptive Response of Mitochondrial Abundance. Circulation 133, 378–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong J et al. , B vitamins attenuate the epigenetic effects of ambient fine particles in a pilot human intervention trial. Proc Natl Acad Sci U S A 114, 3503–3508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim I, Rodriguez-Enriquez S, Lemasters JJ, Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462, 245–253 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou W et al. , Exposure scenario: Another important factor determining the toxic effects of PM2.5 and possibl e mechanisms involved. Environ Pollut 226, 412–425 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Winckelmans E et al. , Transcriptome-wide analyses indicate mitochondrial responses to particulate air pollution exposure. Environ Health 16, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kariya S, Hirano M, Furiya Y, Sugie K, Ueno S, Humanin detected in skeletal muscles of MELAS patients: a possible new therapeutic agent. Acta Neuropathol 109, 367–372 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Nicholls TJ, Minczuk M, In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol 56, 175–181 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Chang L, Xu J, Tie X, Wu J, Impact of the 2015 El Nino event on winter air quality in China. Sci Rep 6, 34275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.L. BL, O. LE, Q. X, T. BM, Analysis of pm2.5 in Córdoba, Argentina under the effects of the El Niño Southern Oscillation. Atmospheric Environment 171, 49–58 (2017). [Google Scholar]
- 65.C. P, Population exposure to hazardous air quality due to the 2015 fires in Equatorial Asia. Scientific Reports 6, 37074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.