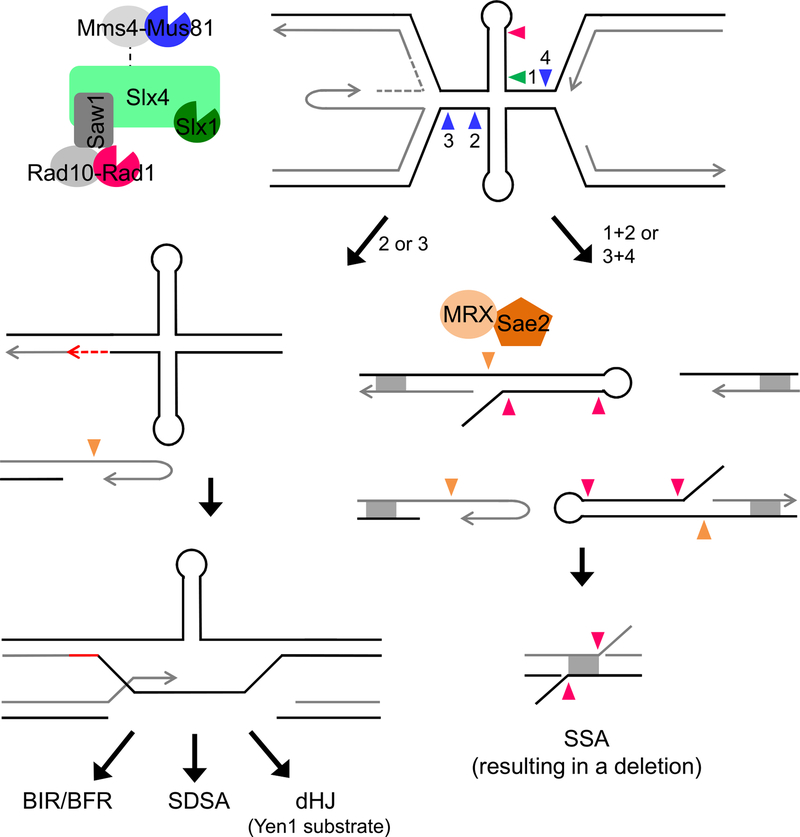
Figure 6. A Model for Mus81 and Slx4 Complex Cleavage of Stalled Fork Substrates Formed by Secondary Structures at Flex1.
Secondary structure forming sequences at Flex1 cause replication fork stalling, which can potentially result in a reversed fork and/or convergence of the approaching fork. A DNA structure and/or the stalled fork is cleaved by SSEs, acting together or sequentially. Cleavages indicated by arrows are color-coded according to the nuclease as depicted. Mus81 cleavage at a stalled fork approaching from the left (arrow 1) will produce a one-ended break (left pathway). The broken end, which may be processed by MRX-Sae2, can invade the intact sister (repaired by gap filling) to initiate repair by homologous recombination, which could proceed by break-induced replication (BIR) or broken fork repair (BFR), synthesis-dependent strand annealing (SDSA), or second-end capture and double Holliday junction resolution by Yen1. Alternatively, if cleavage at stalled forks on either side of the cruciform occurs (arrows 1 and 3) or at the cruciform four-way junction (by coordinated Slx1-Mus81 cleavage, arrows 1 and 2), four ends will be produced. Cleavage of both strands of a single stalled fork will produce three ends, two of which will be hairpin capped if cleavage occurs at the cruciform base. The hairpin-capped ends will be processed by MRX-Sae2. Rad1-Rad10 could also process hairpin loops. Recombinants are recovered by SSA at homologous sequences (DE region of homology denoted by gray box), resulting in deletion of intervening sequences. 3′ non-homologous flaps created during SSA require Rad1-Rad10 and Slx4 nucleases for processing.