Fig. 2. Telomeres and pericentromeres respond differently to Suv39h loss in mESCs.
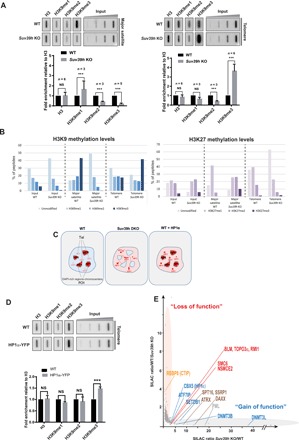
(A) Top: ChIP experiments using antibodies raised against H3 and mono-, di-, and trimethylated H3K9 to monitor H3K9 methylation upon Suv39h loss. Blotted DNA was probed with a telomere-specific probe (right) or a major satellite-specific probe (left). Bottom: Quantifications representing the fold enrichment of H3K9 methylation at major satellites (pericentromere) or telomeres normalized to the total H3 signal and the input relative to wild-type (WT) mESCs. (B) Quantification of relative methylation of histone tails obtained from PICh-purified telomeres and pericentromeres by quantitative MS. H3K9 methylation (left) and H3K27 methylation (right). (C) Model explaining how heterochromatin formation is stimulated by increased HP1α availability at telomeres. Interphase nuclei are represented. Tel, telomere; PCH, pericentromeric heterochromatin organized in chromocenters (DAPI-rich regions). As a result of H3K9me3 erasure at PCH in Suv39h-negative cells, HP1α is delocalized and becomes available to bind to SETDB1-induced heterochromatin, which includes telomeres. Increasing HP1α availability in normal cells by a slight HP1α overexpression results increased heterochromatin formation at telomeres. ***P < 0.005. (D) Left: ChIP experiments using antibodies raised against H3 and mono-, di-, and trimethylated H3K9 in wild-type mESCs overexpressing HP1α–yellow fluorescent protein (YFP). The immunoprecipitated DNA was blotted and probed with a telomere probe. Right: Graph of quantifications of representing the fold enrichment of H3K9 methylation at telomere normalized to total H3 signal and the input relative to wild-type cells. (E) qPICh plots highlighting major protein losses (orange oval) and gains (gray oval) upon SUV39H withdrawal in mESCs. NS, not significant.
