Figure 10.
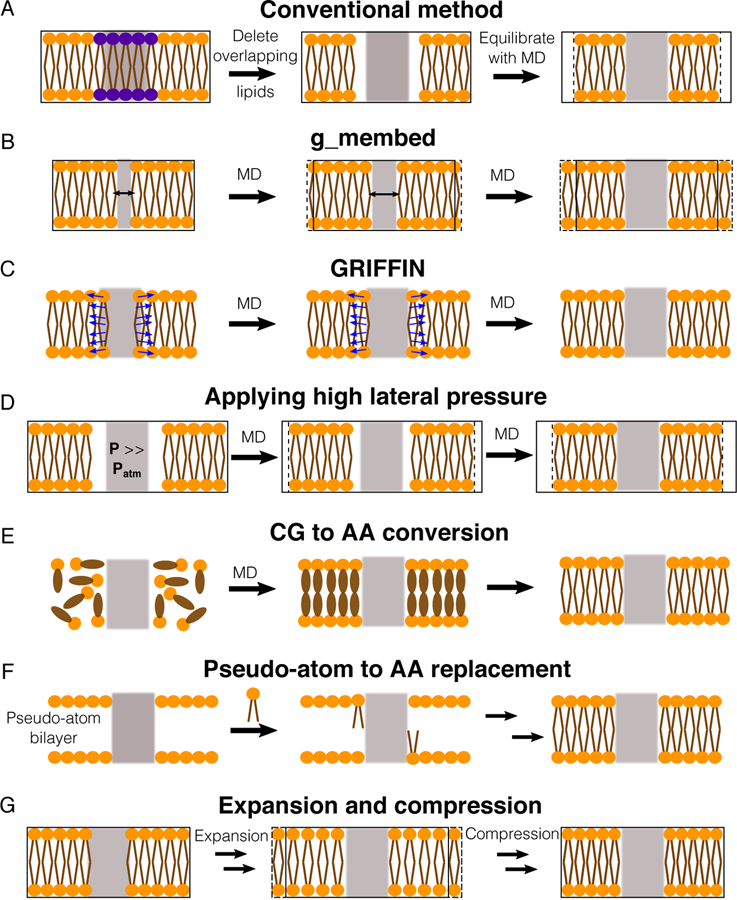
Methods for assembling proteins in membranes. Proteins, lipid head groups and lipid tails are represented by gray rectangles, orange circles and brown lines respectively. Black-solid boxes represent the original dimension of a simulated system, whereas dashed boxes represent changes during the optimization process. (A) The simplest way to optimize lipid packing is to delete lipid molecules colliding the protein and then perform an MD simulation until the system reaches optimal dimensions. (B) g_membed applies a repulsive force to gradually grow the protein to its targeted dimension.201 (C) GRIFFIN applies a repulsive field to carve out lipid molecules inside the protein grid.202 (D) A simulation, in which a high pressure is applied to laterally swallow the protein in the bilayer.203 (E) Flooding simulations can be used to probe lipid binding sites at a CG level first and then transform the assembled complex into an AA model.212 (F) One approach is to first place pseudo atoms or beads of targeted lipid types in a bilayer encompassing the protein according to their cross-sectional areas and then replace the beads with lipid conformations selected from previous MD simulations.72,79,213 (G) Another approach is to perform a series of expansion and compression simulations of the membrane and scaling of lipid size.199
