Figure 4.
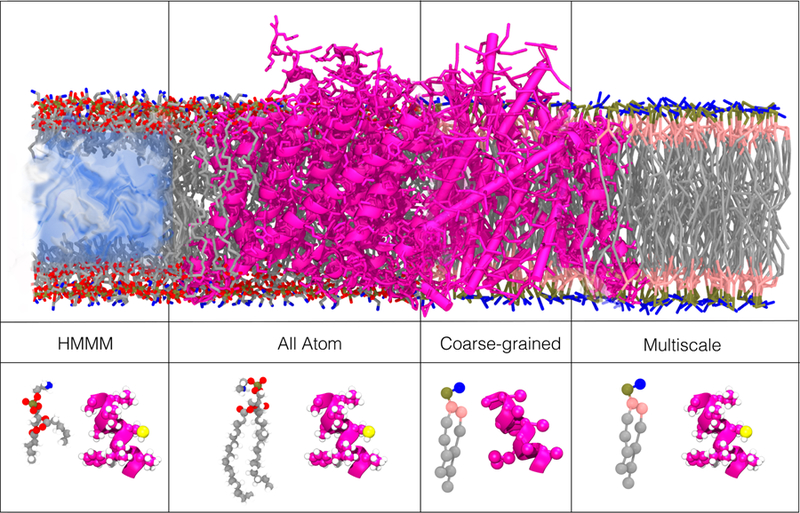
Examples of common resolutions/representations used in the investigation of lipid-protein interactions. The upper panels illustrate different representations for a membrane-embedded ClC channel (PDB:1OTS). The lower panels illustrate each of the representations using a single phosphatidylserine (PS) lipid and a short alpha helix (ClC channel, PDB:1OTS). All-atom (AA) simulations use one interaction-site per atom. In coarse-grained (CG) simulations, several atoms are grouped into one interaction site. Multiscale simulations use a combination of resolutions either in the same simulation or in sequence. The HMMM (highly mobile membrane mimetic) model uses truncated lipids and a membrane core of inorganic solvent to increase lateral diffusion of membrane lipids.
