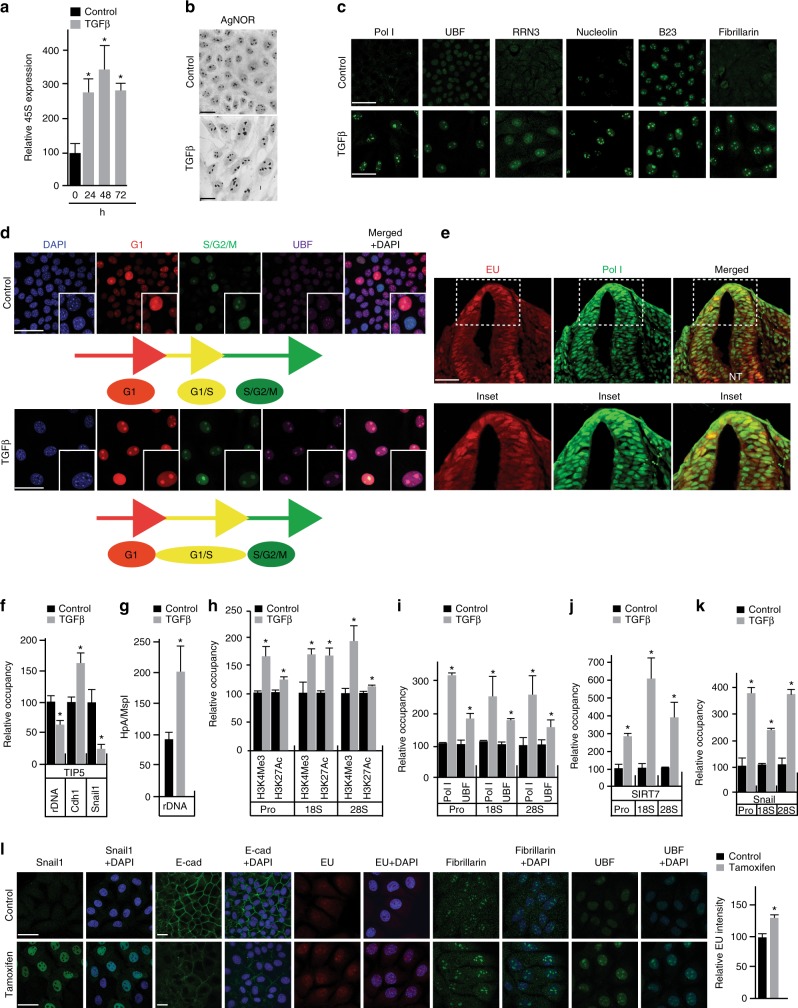
Fig. 2.
Characteristic features of the EMT-associated ribosome biogenesis program. Cell culture experiments in b–k depict proliferating (Control) and TGFβ-treated (TGFβ) NMuMG cells at 48 h assessing changes with treatment. a qRT-PCR of 45S (pre)-rRNA transcript at 24, 48 and 72 hrs (h), P < 0.015. b Silver staining of nucleolar organizer regions (NORs). c Immunostaining of Pol I, UBF, RRN3, Nucleolin, B23, and Fibrillarin (all green). d Cell cycle analysis of proliferating and TGFβ-treated NMuMG cells using the FUCCI technology, DAPI (blue), S/G2/M (geminin, green) and G1 (Cdt1, red), UBF (magenta) and merged. Colocalized green and red fluorescence indicates G1/S arrest (yellow). e rRNA synthesis (EU, red), Pol I (green) and merged (yellow) in the mouse neural crest. NT = neural tube, highlighted by white dotted box. f ChIP analysis of TIP5 binding to the rDNA, E-cadherin (Cdh1) and Snail1 promoters, P < 0.03. g HpaII methylation assay of the rDNA promoter, P < 0.007. h ChIP analysis of H3K4Me3 and H3K27Ac marks to the rDNA promoter, 18S rDNA and 28S rDNA, P < 0.006 and P < 0.005. i ChIP analysis of Pol I and UBF binding to rDNA promoter, 18S rDNA and 28S rDNA, P < 0.015 and P < 0.04. j ChIP analysis of SIRT7 binding to rDNA promoter, 18S rDNA and 28S rDNA, P < 0.005. k ChIP analysis of Snail1 binding to rDNA promoter, 18S rDNA, and 28S rDNA, P < 0.03. l Snail1 (green), E-cad (green), EU (red). Fibrillarin (green), UBF (green) and DAPI (blue) in proliferating (Control) and inducible Snail1 overexpression in NMuMG cells. Quantification of EU, P < 0.0002. All error bars ± SD, n = 3. Asterisk denotes t-test significance. Scale bar for all images = 50 µm